Bioinformatic analysis of anti-phage defense systems in pathogenic strain E. coli O157
Grade 11
Presentation
Problem
Introduction
Escherichia coli O157:H7, also known as verotoxigenic E. coli, verotoxin-producing E. coli, verocytotoxin-producing E. coli, enterohemorrhagic E. coli, and Shiga toxin-producing E. coli (STEC), is a pathogenic strain of toxigenic bacteria, known to cause vomiting, stomach cramps, and severe and occasionally bloody diarrhea.1 Infection can also lead to development of hemolytic uremic syndrome (HUS), a potentially fatal disease that damages blood vessels, thereby damaging the kidneys. Children, especially those under five years of age, immunocompromised individuals, and populations with a genetic prevalence for HUS are more prone to STEC infection.2 E. coli O157 is transferred through fecal-oral transmission by consuming raw or undercooked foods or through person-to-person contact by fecal shedding, the latter of which accounts for approximately 11% of infections.3 In Canada 2019, there were 299 reported cases of E. coli O157:H7, and the rate of infection from 2010 up to that point had been consistently 1.06 cases per 100,000 people.4
Current treatments for patients afflicted with conditions caused by E. coli O157 mostly do not involve hospital admission; however, for more severe cases where the patient develops HUS, treatments may involve kidney dialysis and blood transfusions.1 Studies show that the use of antibiotics in treating E. coli O157 is ineffective and can increase the risk of developing HUS, therefore antibiotics are not utilized to treat E. coli O157. A controlled study conducted by Proulx et al2 on the effect of trimethoprim-sulfamethoxazole (TMP-SMZ) on children with E. coli infections randomized children into a control group and a treatment group, then compared the symptoms. The results showed that the administration of TMP-SMZ did not prevent the onset of HUS. Another study by Ikeda et al5 demonstrated that the use of fosfomycin may decrease risk of HUS; however, almost all the children - all but two - received antibiotic treatment, making the study more of a comparison as opposed to an analysis of fosfomycin singularly. Other studies, by Wong et al6 and Smith et al7 demonstrated that β-lactams and TMP-SMZ increases the risk of HUS development with early administration of treatment in E. coli O157 patients. Due to the lack of a clinical treatment to prevent HUS brought on by E. coli O157, it is important to consider potential alternatives to antibiotics to treat the virus.
Bacteriophages (phages) are viruses that specifically target and infect bacteria such as E. coli O157. Phages infect cells following a lytic or lysogenic pathway. Lytic phages infect bacteria and immediately use bacterial ribosomes and resources to manufacture more phages before lysing the bacteria, while lysogenic phages insert genetic information that can lie dormant in bacteria to be passed through generations before activation, likely due to environmental change.8 The phages used in this study (T1, T4, T5, rv5) are lytic phages. As phages have no observed significant impact on human or animal cells, phage therapy remains a feasible potential treatment option for antibiotic resistant and multidrug resistant bacteria.9
Although the frequency of phage-resistant bacteria is low compared to antibiotics, the issue of potential bacterial mutants as a response to introducing phages as treatment remains. Bacteria may develop phage resistance using several methods. Common defense systems include CRISPR-Cas, restriction-modification (RM) systems, and abortive infection systems. Using the CRISPR-Cas defense system, bacteria integrate invading DNA sequences in repetitive CRISPR sequences, which are transcribed to create a small antisense RNA used by Cas nucleases to cleave invading nucleic acid.10 Restriction modification systems modify specific sequence motifs using a methyltransferase (MTase) so that a cognate restriction endonuclease (REase) will cleave methylated DNA and not host DNA.11
Analysis on bacterial mutations and antiviral defense systems can be done using different bioinformatics pipelines. Whole Genome Sequencing (WGS) is a comprehensive technique for examining genomes and a powerful tool for identifying specific anti-phage defense systems and single-nucleotide polymorphisms (SNPs).12 Other mutations potentially connected to phage resistance or phenotypic alterations can provide insight into evolution of bacteria in response to phage attacks. WGS will be utilized in order to determine anti-phage defense systems and other genetic elements that could contribute to bacterial survival in E. coli O157.
The phages used in this study will be T1 (Rogunavirus), T4 (Tequatrovirus), T5 (Tenquintavirus), and rV5 (Vequintavirus). Previous results have shown that the most effective singular phage out of those used in this study would be T5, a phage associated with reduced E. coli O157 populations;13 however, T5 has demonstrated lower efficacy when working with other phages in a cocktail, particularly with rV5.14 T4 has shown to be the next most effective individually, with T1 being comparable, and rV5 the least effective.
Prolonged exposure to phages may result in phage-resistant bacteria, leading to failure of therapy.14 This remains an obstacle to development of a generalized line of phage therapies. The present study will explore possible solutions to avoid phage resistance, examining the genetic mutations in E. coli O157 that stem from exposure to phages T1, T4, T5, and rv5. Understanding the genetic basis of phage-driven mutations in E. coli O157 can provide insights on evolution of bacteria, contribute to the development of phage therapy, and reveal targets for antimicrobial treatments.
Research Question
What would be the genetic modifications/variations and antiviral defense mechanisms that E. coli O157 develops in response to exposure to phage attacks that contribute to phage resistance?
Method
Bacteria and Phages used in study
In this study, bacteria E. coli O157:H7 was cultured in tryptic soy broth (TSB). Phages (T1, T4, G5, rv5) were used in the study and previously propagated using host bacterial culture.
Phage Exposure Experiments
Previously, E. coli O157 bacterial strain was exposed to the four types of phages (T1, T4, T5, rv5) using the Wright et al. method.15 Phage mutants were created by exposing bacteria to phage and bacterial colonies with abnormal morphology (irregular, star-shaped, or large) were isolated and purified on tryptic soy agar (TSA) plates, followed by confirmation using a microplate assay.
DNA extraction and WGS
A total of 12 phage-resistant bacterial isolates were subjected to DNA extraction using the NucleoSpin DNA extraction kit (Takara Bio, Shiga) and samples were sent to Genome Quebec for sequencing.
The resultant paired-end reads FASTQ files were processed using default settings through Galaxy (https://usegalaxy.org/). FastQC (4.0) was used for quality control on raw reads. The fastp tool was used to trim adapter ends off of sequences and remove low quality reads. Post-trim quality was checked using Phred quality score 20/30, >50% GC content, and >90% good quality reads. Resultant trimmed reads were processed through Shovill using default settings to get the assembled bacterial genomes in FASTA files.
Defense System Analysis
Antiviral Defense Systems were analysed using two pipelines: DefenseFinder (https://defensefinder.mdmlab.fr/) and Prokaryotic Antiviral Defense Locator (PADLOC) (https://padloc.otago.ac.nz/padloc/). Data from both pipelines was sorted and loaded to comma-separated values (CSV) file. R Studio was used for analysis and visualization. Libraries used were ggplot2, reshape2, ggrepel, vegan, pheatmap, and ape. The reference genome (R508 isolate) and experiment control strains were used as reference.
SNP Analysis
FASTA files from Shovill were annotated using Prokka (1.14.5). Resultant .gff files were run through Roary (3.13.0) to analyse gene presence and absence, then Scoary (1.6.16) to relate specific genes with specific traits. Data was sorted into CSV files and .txt files for visualisation using R Studio and Geneious Prime respectively.
Research
Background Research
Link to updated: https://docs.google.com/document/d/13_vT_yBv03Hd7E6ihZlo2GWsK_tM0dga7Mbswa2Mnx0/edit?tab=t.xf6djmm01ep5#heading=h.iifjoxinh8hf
Phage diversity, genomics and phylogeny
08/10/2024
- Abstract
- Massive number and variety of phages in a variety of environments
- Genetically very diverse
- proteins in formation of viral particles are extremely similar
- Introduction
- phages outnumber bacteria in some ecosystems, sometimes by tenfold
- found in every explored biome
- aquatic environment
- major roles in biogeochemical cycling
- short-circuit carbon flow by killing bacteria - viral shunt
- major roles in biogeochemical cycling
- human gut
- mainly in lysogens
- affects physiology and metabolism of hosts.
- mainly in lysogens
- aquatic environment
27/11/2024
- structural morphologies
- tailed phages - double-stranded DNA (dsDNA)
- other morphologies
- non-tailed virions
- single-stranded DNA (ssDNA)
- single-stranded RNA (ssRNA)
- double-stranded DNA (dsDNA)
- due to mosaicism
- large-scale viral metagenomics on recent years
- discovered phages with genome size almost 10times larger than average
- continuous identification of non-tailed dsDNA and ssDNA phages
- many identified share no detectable homology with reference phage genomes
- summary of review
- diversity and four levels of organization
- unique phage morphologies and structural proteins of viruses
- genomic diversity and scarceness in gene similarities
- some proteins still formed with differing genes
- viral diversity at community level
- phage abundance & composition vs ecosystem
- gene exchanges between phages
- mosaicity and drive diversification
- diversity and four levels of organization
- Morphological and structural diversity
- phage genomes
- DNA or RNA
- ds or ss
- packaged into capsid
- polyhedral
- Microviridae, Corticoviridae, Tectiviridae, Leviviridae, Cystoviridae
- filamentous
- Inoviridae
- pleomorphic
- Plasmaviridae
- tail connection
- Caudovirales
- Myoviridae, Siphoviridae, Podoviridae, Ackermannviridae, Herelleviridae
- most phages - tail with dsDNA, belong to Caudovirales order
- Caudovirales share capsid protein (MCP) fold (HK97 fold)
- HK97 identified following X-ray crystallography of capsid of phage HK97
- capsid connected to tail through connector complex
- often a portal protein coupled to head completion or connector proteins
- portal complex is a dodecameric ring with similar overall structure shared between most tailed phages despite low sequence similarity
- capsid completion or connector proteins also form dodecameric rings
- homologues of proteins found in various phages that have contractile and non-contractile tails
- tails of Siphoviridae
- central tape measure protein surrounded by a tail tube and ending with a terminator protein
- similar for Myoviridae phages
- additional layer (protein sheath) enables contraction for insertion of tail tube into host during infection
- capsid-tail joining protein gpFII (Siphoviridae phage lambda) - similar tertiary fold to tail tube protein gpV, adopts same quaternary structure when assembled
- gpV also shares structural homology with tail tube of Myoviridae and components of bacterial type VI secretion system (Hcp1 protein)
- folds of gpFII and gpV similar to baseplate hub of myophages T4 and Mu, no sequence homology
- important building blocks of Siphoviridae and Myoviridae
- Podoviridae (Excherichia coli phage T7)
- short, noncontractile tail
- tube-like extension of tail penetrates membranes of host
- short, noncontractile tail
- Caudovirales
- polyhedral
- Membrane-containing phages
- Phages with small isocahedral capsids or a filamentous morphology
- Two sides of the same coin
- phage genomes
- Genomic diversity and viral metagenomics
- Number of complete genomes
- Range in genome size
- Contribution of viral metagenomics to exploring phage genomic diversity
- Beyond viral metagenomics
- Distribution and abundance
- Phages in marine environments
- Phages in the soil
- Phages in the human gut
- Evolutionary relationships
- Genetic mosaicism as the main actor in phage evolution
- Horizontal gene transfer mechanisms at a glance
- Temperate phages are the brokers in HGT
- Evolutionary relationships between phages differ by host
- A network representation of phage phylogeny
- Conclusions
Revenge of the phages: defeating bacterial defences
09/10/2024
- Abstract
- article about tactics used by bacterial resistance mechanisms in phages
- adsorption inhibition
- restriction-modification
- CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated proteins) systems
- abortive infection
- article about tactics used by bacterial resistance mechanisms in phages
24/10/2024
- Introduction
- bacteriophages
- phages and hosts coexist in state of co-evolutionary equilibrium
- Estimation that phages outnumber bacterial hosts by tenfold
- bacteriophages
24/01/2025
- variety of mechanisms to protect against phages
- inhibition
- cleavage of invading genome
- induction of altruistic cell suicide
- many still succumb to phages
- phages evolved diversified strategies to thrive in bacterial cells
- point mutations
- genome rearrangements
- genomic exchange
17/03/2025
- dynamics of phage-host interactions
- monitoring phage pop to maximize product yield and quality in biotech and food manufacturing
- potential biocontrol agents
- more developed understanding of phage-bacterium co-evolution
- predict and limit phage-resistant bacteria
- more developed understanding of phage-bacterium co-evolution
- understand environmental impact
- biogeochemical cycling
- forms of phage adaptation
- CRISPR-Cas immunity
- anti-CRISPR proteins
- Abi systems
- hijacking host antitoxins to prevent
- review discusses strategies used by phages to overcome bacterial resistance mech
- CRISPR-Cas immunity
- Access to host receptors
- efficient adsorption achieved by targeting surface receptors
- adsorption and injection both necessary for replication
- important to look at interaction between phage receptor binding protein (RBP) and bacterial cell surface receptor to define kinetics
- bacterial receptors
- belong to various biochemical families
- mainly surface proteins, polysaccharides, lipopolysaccharides (LPSs)
- accessibility and distribution on bacteria surface influences efficiency of infection
- bacteria can resist infection by modulating availability (access to receptors)
- phages can regain adsorption ability
- maintains, changes, or expands host range
- phages can regain adsorption ability
- Adapting to new receptors
- Digging for receptors
- Stochastic recognition of variable host receptors
- Battling restriction-modification systems
- Passive mechanisms of phage evasion
- Active mechanisms of phage evasion
- Evading CRISPR-Cas systems
- Evasion by mutation
- Evasion by anti-CRISPR genes
- Phage-encoded CRISPR-Cas systems
- Escaping abortive-infection mechanisms
- Abortive-infection toxin-antitoxin systems
- Conclusions
Resistance Evolution against Phage Combinations Depends on the Timing and Order of Exposure
09/10/2024
- Abstract
- Promising alternative to chemo antibiotics
- lack of understanding of how bacteria evolve resistance against multiple phages
- quantitatively weaker resistance with simultaneous exposure than sequential exposure
- order of phage exposure determined fitness cost of sequential resistance
- some orders impose higher fitness costs than the same phages in diff. order
- suggests that phage combinations can be optimized to limit strength of evolved resistance and maximizing associated fitness cost
- promote long-term efficacy
- suggests that phage combinations can be optimized to limit strength of evolved resistance and maximizing associated fitness cost
- Importance
- rising rates of antibiotic resistance globally
- alternative to antibiotics
- optimization of phage therapy requires understanding of how bacteria evolve resistance against combinations of phages
- use of simple lab experiments and genome sequencing to find timing and order of phage exposure
- determine strength, cost, mutational basis of resistance evolution in Pseudomonas aeruginosa
- Findings suggest that combinations can be optimized to limit emergence and persistence of resistance
- use of simple lab experiments and genome sequencing to find timing and order of phage exposure
- rising rates of antibiotic resistance globally
- Keywords
- Pseudomonas aeruginosa
- bacteriophage therapy
- bacteriophages
- evolutionary biology
- resistance evolution
- Introduction
- evolution of bacterial resistance could limit sustainability of phage-based treatments
- phages initiate infection by binding to molecules on bacterial cell surface
- adsorption
- Common resistance prevents adsorption
- modifies targeted surface molecules that act as phage receptors
- mutations affect regulation or biosynthesis of surface molecule
- potential loss or altered structure of receptor
- due to receptor serving other functions (motility, cell membrane integrity, nutrient transport), alteration of receptor can limit success of mutants in absence of phages
- high fitness cost
- mutations affect regulation or biosynthesis of surface molecule
- modifies targeted surface molecules that act as phage receptors
- resistance against single phage occur at high frequency
- phage kills susceptible genotypes
- resistance mutations spread fast
- due to this, phage therapy involves application of multiple phages to limit emergence of resistance and target wider range of genotype
- resistance to multiple phages unlikely to occur simultaneously
- resistance to multiple phages greatly inhibit bacteria's ability to survive in competition to non-resistant bacteria due to high associated fitness costs
- phage kills susceptible genotypes
- cross-resistance
- most likely to occur if phages target same bacterial surface receptor
- likely that successful combinations of phages target multiple distinct cell surface receptors to limit cross-resistance
- regulators protecting against phages binding distinct receptors are rare and incur far higher fitness costs than those only affecting one receptor
- application of multiple phages can affect trajectory of resistance evolution
- Previous experimental studies suggest sequential can be as effective as simultaneous exposure, but selection for qualitatively different resistance mutations was not tested
- likely that resistances selected under sequential and simultaneous exposure differ
- sequential provides opportunity to accumulate multiple, specific resistance mutations
- simultaneous exposure may be more likely to select for single, generalist cross-resistance mutation
- potential effective treatment
- phage exposure, then antibiotics
- order of exposure important, but effect on evolution of resistance to phage combinations is unclear
- Test described
- evolution, genetic basis, and cost of resistance against pairs of phages targeting same or different cell surface receptors (lipopolysaccharide [LPS] and type IV pilus) in Pseudomonas aeruginosa PAO1 strain
- effect on timing and order effects when phages are simultaneously or sequentially exposed
- sequential exposure allowed accumulation of multiple phage-specific resistances in mutations in cell surface receptor targets of phage adsorption without trade-offs in strength of resistance
- order of phage exposure determined fitness costs (sequential)
- some sequential orders imposed far higher fitness costs than same pair in reverse
- evolution, genetic basis, and cost of resistance against pairs of phages targeting same or different cell surface receptors (lipopolysaccharide [LPS] and type IV pilus) in Pseudomonas aeruginosa PAO1 strain
Escherichia coli (e Coli 0157 H7)
08/10/2024
- produces Shiga toxin
- enterohemorrhagic bacterial strain
- food and waterborne pathogen
- symptoms
- diarrhea
- hemorrhagic colitis
- hemolytic-uremic syndrome (HUS)
- transmission via fecal-oral route post-consumption of contaminated+undercooked liquids, foods, and through fecal shedding
- shiga toxins
- key factor contributing to development of many gastrointestinal illness
- outlines evaluation and treatment of E. coli O157: H7 infection
Efficacy of Individual Bacteriophages Does Not Predict Efficacy of Bacteriophage Cocktails for Control of Escherichia coli O157
24/10/2024
- On an experiment evaluating effectiveness of phages AKFV33 (Tequintavirus, T5) and AHP24 (Rogunavirus, T1), wV7 (Tequatrovirus, T4) and AHP24S (Vequintavirus, rV5) and 11 cocktail combinations of four phages
- Evaluated in vitro for biocontrol of six common phage types of E. coli O157
Why antibiotics should not be used to treat Shiga toxin-producing Escherichia coli infections
28/10/2024
- Introduction
- Shiga toxin (stx)-producing Escherichia coli (STEC)
- can cause hemolytic uremic syndrome (HUS)
- E. coli O157:H7
- typically present with acute, painful bloody diarrhea
- antibiotics bad
- increases development of HUS
- article is a review of cohort studies and meta-analyses that make up current opinion in field
- Shiga toxin (stx)-producing Escherichia coli (STEC)
- Microbial Background
- Table 1
- Shiga toxin (Stx), Stx1, Stx2
- The principal extracellular toxin of Shigella dysenteriae serotype 1. Stx1 is nearly identical to Stx. Stx1 and Stx2 are ~57% identical at the amino acid level. Synonyms: verotoxin (VT), Shiga-like toxin (SLT)
- Shiga toxin-producing E. coli (STEC)
- STEC contain genes encoding one or more variants of Shiga toxin 1 and/or2. Synonyms: verotoxigenic or verocytotoxigenic E. coli (VTEC), enterohemorrhagic E. coli (EHEC), Shiga-like toxin (SLT) -producing E. coli (SLTEC)
- High risk STEC
- An STEC that contains a gene encoding Stx2
- Hemolytic uremic syndrome (HUS)
- a thrombotic microangiopathy characterized by nonimmune hemolysis (HCT <30%), thrombocytopenia (platelet count <150,000mm-3), and azotemia (serum creatinine > upper limit of normal for age.
- Shiga toxin (Stx), Stx1, Stx2
- overarching concern is whether pathogen produces Stx2 due to strong association with HUS ("high-risk")
- almost all produce (~⅔ additionally produce Stx1)
- E. coli O157:H7 is a reasonable indicator of presence of high-risk STEC
- Regional differences
- North America
- STEC of non-E. coli O157:H7 less frequently produce Stx2 or cause bloody diarrhea or HUS
- Continental Europe
- non-O157:H7 STEC more frequently produce Stx2
- same level of HUS risk as E. coli O157:H7
- non-O157:H7 STEC more frequently produce Stx2
- most documented high risk data is on children and O157:H7, likely to be applicable to all high-risk serotypes and patients of all ages
- North America
- Table 1
- Clinical Course and Hemolytic Uremic Syndrome
- high-risk STEC infections usually begin with non-bloody but painful diarrhea which becomes bloody over 2-4 days
- HUS symptoms
- nonimmune hemolytic anemia, thrombocytopenia, azotemia
- most severe complication of STEC
- first described in 1955
- etiology found when Karmali et al. found verotoxigenic (toxic to Vero cells) E. coli in 12/16 children with postdiarrheal HUS
- several case definitions of HUS
- age-focused cut-points to identify HUS
- stringent and specific
- age-focused cut-points to identify HUS
- occurs in 15-20% of childhood E. coli O157:H7 infections
- case definition based on days 5-13 (day 1 is first day of diarrhea)
- 50% or more of children with STEC-associated HUS require renal replacement therapy
- HUS fatality rate is 1-3%
- children under 10 have highest incidence of high risk STEC and developing HUS
- STEC-associated HUS caused by early-in-illness toxemia, injuring vascular endothelium, preceding anemia, thrombocytopenia, azotemia
- Hemolytic Uremic Syndrome and Antibiotics - The First Decade
- Karmali et al. noted that many patients had received various antibiotics at various stages of disease, reiterated in a separate study in Seattle
- strongly suggested antibiotics are not effective
- comparison between STEC patients with and without HUS
- no single study was large enough to demonstrate a lack of efficacy of antibiotics
- Proulx et al only recorded study with randomized, controlled trials for antibiotic effectiveness in reducing HUS likelihood
- randomly allocated 47 children to trimethoprim-sulfamethoxazole (TMP-SMZ) or none
- no significant differences between groups
- enrollment required isolation of E. coli O157:H7, which delayed treatment to median of 7.4 days into illness, where HUS is most commonly diagnosed
- authors recognized children were enrolled late and stressed importance of earlier illness detection in future trials
- cohort studies soon suggested lack of efficacy of antibiotics regardless
- Hemolytic Uremic Syndrome and Antibiotics: 1996 to the Present
- field was influenced by large cohort studies and one meta-analysis
- 1996 E. coli O157:H7 outbreak in Sakai, Japan (Ikeda et al)
- comprehensive study of antibiotic treatment in large scale outbreak related to radish sprouts
- defined HUS rigorously
- only analyzed culture-positive cases
- 14/128 children (10.9%) receiving fosfomycin in the first five days of illness developed HUS vs 30/231 (13.0%) of 231 children who did not receive fosfomycin
- statistically significant differences only identified in children who started fosfomycin on day 2
- led to Infectious Diseases Society of America (IDSA) Guidelines Committee to declare fosfomycin maybe safe and potentially improve clinical course of infections
- a widely cited meta-analysis stated that administering fosfomycin within the first 2 days of illness was associated with a significantly reduced risk of HUS
- improbable that it would only be effective for 2nd day and not rest
- all but 2 children in study received antibiotics
- does not demonstrate superiority
- Prospective cohort studies
- Wong et al demonstrated β-lactams and TMP-SMZ increase risk of developing HUS when administered early in infection
- after adjusting for white count, presence of bloody diarrhea, pain med. use, age, vomiting (severity) no evidence that antibiotic use influenced by severity of illness
- Wong et al demonstrated β-lactams and TMP-SMZ increase risk of developing HUS when administered early in infection
- Karmali et al. noted that many patients had received various antibiotics at various stages of disease, reiterated in a separate study in Seattle
Hemolytic uremic syndrome associated with Escherichia coli O157:H7 infection in older adults: a case report and review of the literature
10/10/2024
- Abstract
- HUS is associated with STEC
- acute renal failure in children
- sporadic STEC HUS cases in adults in the USA
- mortality rates highest in patients >60
- 86 year old Caucasian woman
- treated for suspected thrombotic thrombocytopenic purpura, worsening neuro and renal functions despite plasmapheresis
- normal ADAMTS13 and positive stool confirmed E. coli O157 and STEC HUS
- shifted to aggressive supportive care
- HUS caused death despite conventional treatment
- Importance of recognising STEC HUS as etiology of microangiopathic hemolytic anemia in older people
- supportive care is current best approach to STEC HUS
- plasmapheresis and eculizumab (complement inhibitor) shown ineffective in STEC HUS
- HUS is associated with STEC
- Background
- Microangiopathic hemolytic anemia (MAHA)
- effects
- erythrocyte fragmentation
- elevated lactate dehydrogenase (LDH)
- microvascular occlusion
- presenting with normal ADAMTS13 consists of disease spectrum including HUS
- effects
- HUS
- classical or typical HUS associated with STEC or invasive pneumococcal infection
- atypical HUS (aHUS) unrelated to STEC driven by complement activation and dysregulation
- one of main causes of acute renal failure in children
- supportive care and dialysis current mainstream therapy
- not many cases in adults worldwide (reported)
- outbreak of HUS with STEC O104:H4, Germany, 2011
- highest rate of mortality in adults >60
- rarity of adult presentation results in delayed diagnosis and limited treatment options
- 86 year old woman with STEC-HUS
- differentiate from other MAHA entities
- Microangiopathic hemolytic anemia (MAHA)
- Case presentation
- transferred to intensive care from another facility for higher level of care for thrombotic microangiopathy and acute renal failure
- symptoms
- presented to emergency department with acute onset of abdominal pain and bright red color per rectum
- reported breathing difficulty
- CT found nonspecific results suggestive of colitis, therapy started with levofloxacin and metronidazole
- no features of UTI in urine
- difficulty of speech
- word-finding
- slurring
- family-reported
- no other neurological deficits
- intubated at other hospital
- altered mental status
- need for supplemental oxygen prior to transfer
- diagnosed of MAHA with suspicion of thrombotic thrombocytopenic purpura (TTP) prior to transfer
- continued metronidazole, cefepime, tigecycline after transfer
- comorbities of essential hypertension, hyperlipidemia, hypothyroidism
- no reported history of infectious, autoimmune, or hematologic disease
- examination
- skin findings of purpura and ecchymoses were noted
- lab data
- elevated creatinine
- markedly elevated LDH
- markedly reduced haptoglobin
- serum ferratin was 739.8 ng/mL
- transferrin sat was 42%
- cobalamin and folate within normal
- peripheral smear
- thrombocytopenia and MAHA features
- approached as MAHA with high suspicion for TTP
- STEC-HUS in differential due to bloody diarrhea
- started PLEX beside methylprednisolone 1mg/kg twice daily waiting for ADAMTS13 and Shiga-toxin assays
- also underwent hemodialysis for acute renal failure
- initial improvement in hemolysis and thrombocytopenia
- lab and clinic parameters of MAHA failed to improve in subsequent hospitalization days
- ADAMTS13 activity reported as normal
- 92%, normal is 68-163
- stool sample positive for STEC
- consistent with diagnosis of STEC-HUS
- discontinued corticosteroid and plasmapheresis
- discussed treatment options
- family decided on palliative measures
- passed on fourth day of hospitalization
- Discussion
- STEC-HUS
- characterized by MAHA, thrombocytopenia, acute renal injury (6-9% of STEC infections)
- neurological involvement also associated, less common
- pathophysiology
- organ damage through vasculoendothelial injury
- Stx
- released by toxic strains of bacteria released into gastrointestinal tract
- absorbed into systemic circulation binding to globotriaosylceramide (Gb3) on surface of vascular endothelial cells in different target organs
- kidney, brain
- absorbed into systemic circulation binding to globotriaosylceramide (Gb3) on surface of vascular endothelial cells in different target organs
- injures endothelial cells
- inhibiting protein synthesis, inducing broad inflammatory response, releasing cytokines and chemokines
- increases risk of thrombosis and organ damage
- activates alternative pathway of complement system by binding to factor H proteins, (complement control systems) and reducing cell surface activity
- Stahl et al studied effects of STEC on complement system and illustrated increased binding of microparticles from platelets and monocytes in plasma to C3 and C9 factors
- released by toxic strains of bacteria released into gastrointestinal tract
- Presentation
- prodromal illness with abdominal pain, vomiting, diarrhea preceding MAHA, thrombocytopenia, acute kidney injury (AKI) by 5 to 10 days
- MAHA
- characterized by hemoglobin levels <8g/dL, negative Coombs test, elevated serum indirect bilirubin concentration, elevated serum LDH level, decreased serum haptoglobin concentration, peripheral blood smear with large number of schistocytes and reticulocytes and nucleated red blood cells
- Thrombocytopenia
- characterized by platelet count below 150000/mm^3
- AKI
- hard to define due to varying severity of renal involvement
- hematuria and proteinuria to severe AKI and oliguria requiring dialysis
- 50%of HUS patients require dialysis in acute phase
- hard to define due to varying severity of renal involvement
- additional organ involvement
- 20-25% of patients develop life-threatening neurological involvement with severe neurological symptoms
- lethargy, apnea, coma, seizures, cortical blindness, hemiparesis
- magnetic resonance imaging in brain in patients with severe neurological involvement
- abnormal findings in basal ganglia, thalami, brainstem
- severe central nervous system (CNS) involvement associated with higher mortality
- 20-25% of patients develop life-threatening neurological involvement with severe neurological symptoms
- studies focusing on adults
- Gould et al
- Foodborne Diseases Active Surveillance Network (Foodnet)
- population-based surveillance for O157 infection and HUS from 2000-2006 in eight different US states to explore demographic risk factors for development of STEC-HUS and evaluation of mortality rates by age
- young females under 5 had highest HUS development risk following O157 infection, while people over 60 had highest mortality rate with or without HUS
- Foodborne Diseases Active Surveillance Network (Foodnet)
- German study
- neurological symptoms
- higher in adults than children
- higher mortality rates
- deaths occuring acutely during initial hospitalization mainly due to cerebral edema and infarction, sepsis, electrolyte disturbance
- neurological symptoms
- Gould et al
- treatment modalities
- antithrombotic agents, PLEX, tissue-type plasminogen activator, oral Stx-binding agent, eculizumab included in range studied to manage STEC-HUS
- main focus on PLEX and eculizumab: most frequently studied therapies
- PLEX
- removal of Stx-like toxin and prothrombotic factors caused by toxin and inflammatory mediators
- replace with coagulation, tissue, complement factors
- 2010 guidelines of American Society for Apheresis
- listed as one recommended treatment for STEC-HUS
- German study
- proposed general supportive care has same outcome as PLEX clinically
- Initial case presentation
- led to initial improvement in hemolysis and consumptive thrombocytopenia
- should be initiated early when MAHA is suspected
- difficult to differentiate HUS and TTP
- time lag for confirmation
- removal of Stx-like toxin and prothrombotic factors caused by toxin and inflammatory mediators
- eculizumab
- antithrombotic agents, PLEX, tissue-type plasminogen activator, oral Stx-binding agent, eculizumab included in range studied to manage STEC-HUS
- STEC-HUS
Adapting to new threats: the generation of memory by CRISPR-Cas immune systems
10/10/2024
- Summary
- Clustered, regularly interspaced, short palindromic repeats (CRISPR)
- CRISPR loci and associated genes confer bacteria and archaea with adaptive immunity against phages and other invading genetic elements
- memory of past infections to deal with recurrent infections efficiently is a fundamental requirement of any immune system
- CRISPR-Cas adaptation relies of ability to memorize invading DNA sequences and integrate them in repetitive sequences of CRISPR in form of spacers
- transcription generates a small antisense RNA used by RNA-guided Cas nucleases to cleave invading nucleic acid to prevent infection
- acquisition of new spacers allows rapid adaptation
- CRISPR-Cas adaptation relies of ability to memorize invading DNA sequences and integrate them in repetitive sequences of CRISPR in form of spacers
- recent studies begun elucidation of genetic requirements for adaptation
- non-stochastic
- selection of spacers influenced by several factors
- review of current knowledge of CRISPR adaptation mechanism
- Clustered, regularly interspaced, short palindromic repeats (CRISPR)
- Introduction
- bacteria and archaea evolved to thrive in hostile environments under constant phage threat
- numerous strategies to prevent phage infection
- abortive infection, surface exclusion, restriction modification
- highly effective, but non-specific immunity
- CRISPR-Cas is an adaptive defence mechanism against mobile genetic elements
- numerous strategies to prevent phage infection
- History
- discovery of E. coli in 1987
- widespread among bacteria and archaea
- CRISPR-associated (cas) genes
- adjacent to array of repeat and spacer sequences
- encode a diverse family of Cas proteins carrying predicted functional domains of proteins
- participate in nucleic acid transactions, such as DNA-binding proteins, nucleases, polymerases, helicases
- CRISPR array
- identical non-adjacent sequences interspaced by similarly sized variable sequences (repeats and spacers)
- AT-rich leader sequence located upstream of first promotes transcription of CRISPR array
- essential for spacer acquisition
- repeats usually conserved within same locus, most cases contain partially palindromic sequences
- spacers are highly diverse even among closely related strains
- initially exploited for strain typing purposes
- 3 independent bioinformatics studies revealed homology between spacer sequences and mobile genetic elements
- led to hypothesis that CRISPR may provide protection against phages and plasmids
- confirmed soon after to provide sequence-specific interference against all prokaryotic routes of horizontal gene transfer
- bacteriophage infection
- plasmid conjugation
- transformation
- 3-step defence pathway
- fragment of invading nucleic acid (protospacer) incorporated into CRISPR array with synthesis of additional repeat unit
- adaptation
- responsible for adaptive features of CRISPR
- crRNA biogenesis phage
- CRISPR locus transcribed and processed into mature guide RNA (crRNA)
- crRNA recruits effector complexes
- guides to target by base pairing with invading nucleic acid
- interference
- cleal=vage of exogenous genetic element
- 3 types of CRISPR systems (I-III) with several subtypes depending on gene comp and architecture of respective cas operons despite similar pathway
- CRISPR adaptation (immunization or spacer acquisition) poorly understood
- fragment of invading nucleic acid (protospacer) incorporated into CRISPR array with synthesis of additional repeat unit
- spacer acquisition
- first demonstrated under lab conditions in 2007 for type II-A system of Streptococcus thermophilus
- examined CRISPR array of phage-immunized bacteria
- addition of new repeat-spacer units with new spacers perfectly matching sections of genome with challenging phage
- Mutants with shared sequence spacers were resistant to both viruses
- established CRISPR-Cas as adaptive, sequence-specific immune system
- corroborated in other bacteria and archaea
- first demonstrated under lab conditions in 2007 for type II-A system of Streptococcus thermophilus
- bacteria and archaea evolved to thrive in hostile environments under constant phage threat
- Target requirements: can any sequence of the invader's genome become a CRISPR spacer?
- Investigation
- aligned newly acquired spacers from S. thermophilus system looking for motifs
- conserved sequence outside target (protospacer)
- Protospacer Adjacent Motif(PAM)
- suggested adaptation machinery only requires spacers with adjacent PAMs
- conserved sequence outside target (protospacer)
- aligned newly acquired spacers from S. thermophilus system looking for motifs
- PAM required for interference phase of CRISPR immunity
- PAM mutations in types I and II prevent Cas nuclease cleavage
- Regularly exploited by phages
- avoid CRISPR immunity by mutating PAM sequence
- fundamental to avoid autoimmunity
- CRISPR immunity relies on base-pair interactions between crRNA and target DNA, spacer sequence on CRISPR array would be crRNA target as well
- flanking sequences of a spacer in CRISPR array are repeats lacking PAM, auto-immunity is prevented and only protospacers flanked with correct PAM can be cleaved
- exception of Type III systems as PAM is not evident nor required for cleavage
- TypeIII systems developed different mechanism to prevent auto-immunity
- other mechanisms influencing protospacer choice
- spacer acquisition studies reported unequal distributions of protospacers across various targets
- CRISPR array expansion from S. thermophilus monitored using DNA deep-sequencing upon infection with a lytic phage
- roughly half a million phage-derived spacer sequences analyzed
- top 10% most overrepresented spacers accounted for 99% of identified sequences
- some candidate protospacers that could be theoretically acquired from target based on PAM compatibility were never sampled
- spacer acquisition studies reported unequal distributions of protospacers across various targets
- partial sequence similarities between some endogenous spacers and target, Paez-Espino et al propose priming as possible explanation for strong overrepresentation of certain protospacers
- similar study in type I-E
- all potential protospacer sequences adjacent to PAM used as spacer donors
- highly unequal frequencies
- no correlation observed between protospacer incorporation frequency and nucleotide sequence, melting temperature, GC content, ssDNA secondary structure, transcription pattern
- other investigators detected additional sequence motifs besides PAM influencing acquisition efficiency of protospacer
- all potential protospacer sequences adjacent to PAM used as spacer donors
- Exchanging nucleotide blocks of various lengths upstream or downstream of one high and one low-acquisition protospacer, investigators could reverse rates of acquisition
- suggests DNA motifs at both ends of protospacer responsible for frequent incorporation
- In addition to PAM, a dinucleotide AA motif
- Acquisition Affecting Motif (AAM)
- located at 3' end of protospacer
- can boost rate of incorporation of a protospacer
- over 2 million spacers confirmed overrepresentation of AA motif in highly sampled protospacers
- AAM not present in all highly sampled protospacers
- other DNA motifs may influence sampling frequencies
- different study did not confirm AAM among plasmid-derived spacers
- Other observations
- unknown mechanisms influence spacer choice
- experiment with overexpressed Cas1 and Cas2 in absence of interference machinery
- Yosef et al observed a 200fold preferential acquisition of spacers matching Cas1+2 overexpression plasmid over chromosome
- mechanisms might exist to limit acquisition of self-targeting spacers
- hypothesis unsupported by recent study where preferential acquisition of plasmid-derived protospacers was also observed, but only from plasmid carrying cas genes and not from another reported plasmid in cells
- Yosef et al observed a 200fold preferential acquisition of spacers matching Cas1+2 overexpression plasmid over chromosome
- suggest Cas1 and Cas2 preferentially capture protospacers in close proximity to coding region
- Investigation
- The machinery responsible for spacer acquisition: naive versus primed adaptation
- Cas proteins
- only proteins universally conserved across all types and subtypes of CRISPR-Cas systems are Cas1 and Cas2
- initial studies revealed mutations or deletions of Cas1 or 2 did not imp[act interference and cfRNA maturation in all types
- led to hypothesis that two universal Cas proteins may be involved in adaptation
- in E. coli, overexpression of cas1 and cas2 in absence of other cas genes was sufficient to acquire new plasmid or host-derived spacers
- Yosef et al also found Cas1/2 mediate preferential acquisition of spacers with a correct PAM, demonstrating a mechanism to select spacer sequences flanking this motif as opposed to random acquisition of DNA sequences followed by the selection of those with correct PAMS
- biochemical properties of Cas1 support its role in spacer acquisition
- Pseudomonas aeruginosa Cas1 shown to bind dsDNA in a sequence-independent manner with high affinity
- works as metal-dependent endonuclease which cleaves dsDNA into short fragments, might serve as precursors for new spacers
- remains unclear whether new spacers are cut or copied from invading molecule
- Cas1 can also resolve Holliday junctions and promote DNA integration and recombination events
- could promote integration of spacer sequences into array
- many Cas2 crystal structures have been solved and studied biochemically
- consensus regarding activity unreached
- Cas1 can also resolve Holliday junctions and promote DNA integration and recombination events
- mechanism of acquisition does not require presence of other spacers
- Cas1/2 mediated acquisition can add repeat -spacer units to minimal CRISPR locus consisting of only one repeat sequence
- mech of acquisition does not require other spacers' presence
- (previous exposure to same or related phages)
- "naive" acquisition
- "primed" acquisition would be where spacers have full or partial match
- previous encounter
- mech of acquisition does not require other spacers' presence
- Previously studied primed acquisition
- CRISPR-Cas system harbors genes encoding Cascade (CRISPR associated complex for antiviral defense)
- Cascade complex contains crRNA and is responsible for target recognition
- Cas3 nuclease is responsible for target cleavage
- in addition to Cas1 and 2
- Cas3 nuclease is responsible for target cleavage
- Study that showed
- Cas1/2 mediated acquisition can add repeat -spacer units to minimal CRISPR locus consisting of only one repeat sequence
- Cas proteins
- Insertion of new spacers into the CRISPR array
- Concluding remarks
A mobile restriction-modification system provides phage defence and resolves an epigenetic conflict with an antagonistic endonuclease
03/02/2025
- Abstract
- Epigenetic DNA methylation influences gene expression and allows discrimination between self-DNA and intruders such as phages and plasmids in bacteria
- RM systems use a methyltransferase (MTase) to mod a specific sequence motif
- protects host DNA from cleavage by a cognate restriction endonuclease (REase)
- leaves invading DNA vulnerable
- other REases
- occur solitarily
- cleave methylated DNA
- RM systems and REases are frequently mobile
- influence horizontal gene transfer by altering compatibility of host for foreign DNA uptake
- obscure whether mobile defence systems affect pre-existing host defences
- Study
- epigenetic conflict between RM system (PcaRCI) and methylation-dependent REase(PcaRCII) in plant pathogen Pectobacterium carotovorum RC5297
- PcaRCI RM system provides protection against unmethylated plasmids and phages
- methylation-dependent PcaRCII targets methylation motifs, including the motifs modified by PcaRCI
- co-existence enabled through epigenetic silencing of PcaRCII-encoding gene via promoter methylation by PcaRCI MTase
- comparative genome analyses
- PcaRCII encoding gene was already present and was silenced upon PcaRCI system establishment
- Graphical Abstract
- Introduction
- epigenetic modifications
- heritable but do not change base DNA sequence
- governs diverse processes in higher organisms such as development or emergence of disease
- epigenetics key in bacteria
- most common mark is DNA methylation, catalysed by MTases
- known to occur at adenine residues
- N6-methyladenine (m6A)
- and cytosine residues
- N4-methylcytosine (4mC)
- known to occur at adenine residues
- most common mark is DNA methylation, catalysed by MTases
- MTases may be solitary enzymes
- DNA adenine MTase (Dam) & DNA cytosine MTase (Dcm) in E. coli
- cell-regulating-cycle MTase (CcrM) in Caulobacter crescentus
- adenine MTase A (CamA) in Clostridioides difficile
- can fulfill variety of roles in DNA replication, mismatch repair, cell cycle progression, stress response, pathogenesis
- MTases may occur in association with restriction endonucleases (REases), forming RM systems
- some RM MTases are also known to affect cellular transcriptome
- major function of RM systems is protection against invaders like phages and plasmids
- MTases of RM systems methylate specific base within target motif
- REase cleaves DNA after recognition of same unmethylated sequence
- bacterial genome is protected from cleavage, while unmethylated intruders are degraded by REase
- types of RM systems
- I
- epigenetic modifications
- Materials and Methods
- Bacterial strains and growth conditions
- DNA isolation and manipulation
- DNA and protein sequence analyses
- Preparation of electrocompetent P. carotovorum cells and electroporation
- Transformation assay
- Conjugation efficiency assay
- Bacteriophage isolation and titration
- Extraction of genomic DNA
- Whole-genome sequencing
- RNA extraction and sequencing
- RNA sequencing analysis
- Reporter assay
- Competition assay
- Results
- Pectobacterium carotovorum RC5297 discriminates self and foreign DNA
- A restriction-modification system inhibits plasmid acquisition and phage infection
- Genome methylation by M.Pcal inhibits an alternative defence mechanism
- A methylation-dependent HNH endonuclease provides defence in Pca^deltaRM
- Methylation by M.Pcal represses the pcallR promoter
- The RM system is sparse in P. carotovorum strains and is a part of a variable genomic region
- Co-existence of pcaland pcall loci does not have a significant fitness cost
- Discussion
- Data Availability
The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair
10/10/2024
- Abstract
- abortive infection (Abi) systems included in list of resistance strategies of bacteria
- promote cell death, limit phage replication within bacterial population
- Abi Erwinia carotovora sub atroseptica designated ToxIN
- also functions as toxin-antitoxin (TA) pair, ToxN inhibits bacterial growth and ToxI RNA antitoxin counteracts toxicity
- TA modules currently in 2 classes
- protein
- RNA antisense
- ToxIN defines a new class
- provides viral resistance in a range of bacterial genera against multiple phages
- first demo of novel mechanistic class of TA systems and Abi system functioning in diff. bacterial genera
- Bacteria become infected at 10^25 per second by phages
- outnumbered by phages 10 to one
- facilitated coevolution (evolutionary clash)
- protection
- surface alterations
- prevention of phage DNA injection
- restriction of incoming DNA
- phage-specific immunity through CRISPR
- Abi
- promotes altruistic suicide of infected bacteria
- "self-lysing?"
- majority found on plasmids of Gram-positive lactococcal strains
- some found in Gram-negative species
- E. coli, Vibrio cholerae, Shigella dysenteriae
- often highly toxic after activation
- varied targets
- can act on central cellular processes to inhibit
- replication, transcription of DNA
- protein synthesis
- specific effects
- premature cell lysis AbiZ
- interference of a phage RuvC-like endonuclease by AbiD1
- promotes altruistic suicide of infected bacteria
- toxic protein roles
- recent influx of genomic info
- frequent ID of multiple TA loci on bacterial and archaeal chromosomes
- originally identified as plasmid addiction systems
- widespread nature of TA operons led to discussion of biological role
- TA systems rely on dual activity of toxin and antagonistic antitoxin
- antitoxins are labile compared with their toxins
- when production of both is inhibited, toxin is prioritized to take effect
- toxins of known TA systems, like Abi proteins, can target central cellular processes
- DNA replication & translation
- DNA gyrase (ccd and parDE loci)
- mRNA degradation (relBE and mazEF loci)
- DNA replication & translation
- recent influx of genomic info
- abortive infection (Abi) systems included in list of resistance strategies of bacteria
- Results
- An Eca Plasmid Provides Phage Resistance
- toxIN Encodes an Abi System
- ToxN is a Toxin in E. coli
- toxIN Is Bicistronic
- toxI Encodes an Antitoxin
- ToxIN Is a Reversible Bacteriostatic TA System
- ToxIN Is a Novel Protein-RNA TA System
- ToxIN Defines a New TA Family
- toxIN Acts as an Abi System in Multiple Enteric Bacteria
- Discussion
- Materials and Methods
- Bacterial Strains, Plasmids, and Culture Conditions
- DNA Manipulations and Sequence Analysis
- Subcloning and Sequencing of pECA1039
- In Vitro Mutagenesis of pECA 1039
- Phage Techniques
- ToxN Toxicity Assays
- ToxIN Bacteriostatic, Protection, and Stability Assays
- Mapping the Transcriptional Start of toxIN
- toxIN Promoter lacZ Fusion Experiments
- Construction of ToxI Expression Vectors
- Western Blot Analysis of ToxN
- Construction of Bacillus thuringiensis toxIN Plasmids
Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types
10/10/2024
- Abstract
- Introduction
- Materials and Methods
- PADLOC implementation
- Building HMMs, system classifications and benchmarking
- Results
- Discussion
National Enteric Surveillance Program
18/02/2024
- Overview
- NESP is a program by the Public Health Agency of Canada (PHAC) and the Provincial Public Health Laboratories
- weekly analysis and reporting conducted for 14 different organisms
- cause enteric illness
- 10 are nationally notifiable
- data in surveillance system
- detection of multi-provincial clusters and outbreaks
- guides public health interventions
- designed to integrate with national/international efforts limiting transmission of enteric diseases
- isolates
- 15,142 reported, decrease in previous 5 years (15,541)
- incidence rate of STEC O157 stable since 2010
- 1.06 cases/100,000 in 2019
- increase in incidence rate of non-O157 observed in 2019 (2.5/100,000)
- third year with higher non-O157 higher than O157 report
- Escherichia coli
- current rate has been consistently 1.06 cases/100,000 since 2010
- reported higher than national average in 2019
- Alberta - 2.17 cases
- Manitoba - 1.83 cases
- Newfoundland - 2.49 cases
- Data
- 299 cases of E. coli O157:H7 in 2019
Alberta Public Health Disease Management Guidelines (Haemolytic Uremic Syndrome)
(Alberta public health disease managem...)
18/02/2024
- Epidemiology
- Etiology
- D+ HUS is diarrhea associated, D-HUS is atypical
- 90%+ thought to be caused by VTEC in North America
- Etiology
Interactions of Bacteriophages with Animal and Human Organisms-Safety Issues in the Light of Phage Therapy
23/02/2024
- Abstract
- used to be considered neutral to animals due to lack of phage receptors
- new evidence suggests ability to interact with eukaryotic cells
- significant influence on functions of mammal cells
- summary of interactions
- modulations of cancer cells
- direct and indirect effects
- interactions are robust and should be taken into consideration
- Introduction
- necessity of alternative treatments to multi-drug-resistant bacteria
- phage therapy
- specificity to selected bacteria
- widely regarded as safe
- current literature lacks significant adverse effects
- technically interactions are possible
- interactions
- Ways for Penetration of Animal and Human Tissues by Bacteriophages
- The Epithelial Barrier
- The Circulatory System
- The Endothelial Barrier
- The Blood-Brain Barrier
- The Skin
- Interactions of Bacteriophages with Mammalian Immune System
- Antiphage Innate Immune Response
- Dendritic Cells
- Monocytes and Macrophages
- Granulocytes
- Foreign Particles vs. Innate Immune Response
- Clearance of Bacteriophages
- Phage-Induced Cytokine Response
- Phages as Factors Increasing Bacterial Phagocytosis
- Antiphage Adaptive Immune Response
- Phage Therapy against Bacterial Infections of Lungs
- Antiphage Innate Immune Response
- Interactions of Bacteriophages with the Respiratory System
Bacteriophages (StatPearls)
23/02/2024
- Introduction
- general
- phages
- viruses that infect and replicate solely bacteria
- ubiquitous in environment
- most abundant biological agent
- diverse in size morphology and genomic organization
- composition
- nucleic acid genome
- encased in phage-encoded capsid proteins
- protect genetic material
- mediate delivery to next host
- encased in phage-encoded capsid proteins
- nucleic acid genome
- detailed visualization of hundreds of types
- heads, legs, tails sometimes occur as characteristics
- non-motile - depend on Brownian motion
- phages
- Lytic Replication
- attach
- introduce genome to cytoplasm
- uses host ribosomes to manufacture phage proteins
- host cells rapidly converted to viral genomes and capsid proteins
- assemble into multiple copies of original phage
- host cell passively or actively lysed to release phage
- Lysogenic Replication
- first 2 steps same
- phage genome integrated to chromosome or maintained as episomal
- replicated and passed to daughter cells
- integrated phage genome - prophage
- host bacteria - lysogens
- prophages can convert to lytic replication cycle
- usually in response to environmental change
- general
- Function
- general
- Generalized Transduction
- Specialized Transduction
- general
- Clinical Significance
- Enhancing Healthcare Team Outcomes
- Antibiotic Resistance and Infection Control
- Bacterial Toxin Mediated Disease
Phage-host interplay: examples from tailed phages and Gram-negative bacterial pathogens
(Chaturongakul and Ounjai 2014)
23/02/2024
- Abstract
- phage interactions shape environmental microbial communities
- transduction
- modification of bacterial gene expression patterns
- survival depends on infection
- entry
- coevolution of tail fibers and bacterial receptors determine many infection parameters
- review
- summarizes knowledge between tailed bacteriophages and bacterial pathogens
- phage interactions shape environmental microbial communities
- Introduction
- phages are the most abundant and diversified microorganisms on Earth
- obligate bacterial predators
- can be found everywhere bacteria is
- evolutionary success
- about 20 more successful abundance than bacteria
- phages and bacteria in a constant state of coevolution
- focus of review on physical interactions between tailed phages and Gram-negative bacterial pathogens
- Physical Interactions between Tailed Phages and their Hosts
- Phage-Bacteria: Genetic Give-and-Take
- Phages in Microbial Ecosystems
- New Research Approaches in Phage-Host Interaction
- Conclusions and Perspectives
Clinical Analysis of Whole Genome Sequencing in Cancer Patients
24/02/2024
- Abstract
- Purpose of Review
- Recent Findings
- Summary
- Introduction
- Next-Generation Sequencing and the Cancer Genome
- Changes to NHS Tissue Handling for Genomic Testing
- WGS Variant Calling and Interpretation
- in WGS, bioinformatic algorithms create reports with relevant clinically actionable information
- pipeline
- The Genomics Tumour Advisory Board
- Analysing the WGS Report
- Clinical information
- Quality control
- Somatic variants
- Germline variants
- Tumour mutational burden
- Mutational signatures
- Circos plot
- Outcome
- Genomic Testing and Clinical Trials
- Tumour Heterogeneity and Liquid Biopsy
- The NHS Genomic Medicine Service
- Multi-omics - The Next Step in Cancer Genomics
- Conclusions
A Simple Practical Guide to Genomic Diagnostics in a Pediatric Setting
24/02/2024
- Abstract
- Genomic Testing Strategies
- Whole Genome Sequencing
- sequences protein coding regions and genome
- functional impact of variation outside of exome thus far not established
- many well-established genes have pathogenic variants outside coding sequences
- intronic variants
- upstream variants
- most labs attempt to catch these anyway
- WGS is able to capture structural rearrangements
- CNVs, translocations, inversions
- often embedded in intronic noncoding regions
- likely to replace many other tools
- need to clinically validate ability of tools to replace tools to justify cost and data storage issues
- Whole Genome Sequencing
fastp: an ultra-fast all-in-one FASTQ preprocessor
26/02/2024
- Introduction
- Materials and methods
- Overall design
- Adapter trimming
- overlaps pairs and trims adapters outside of pair
Data
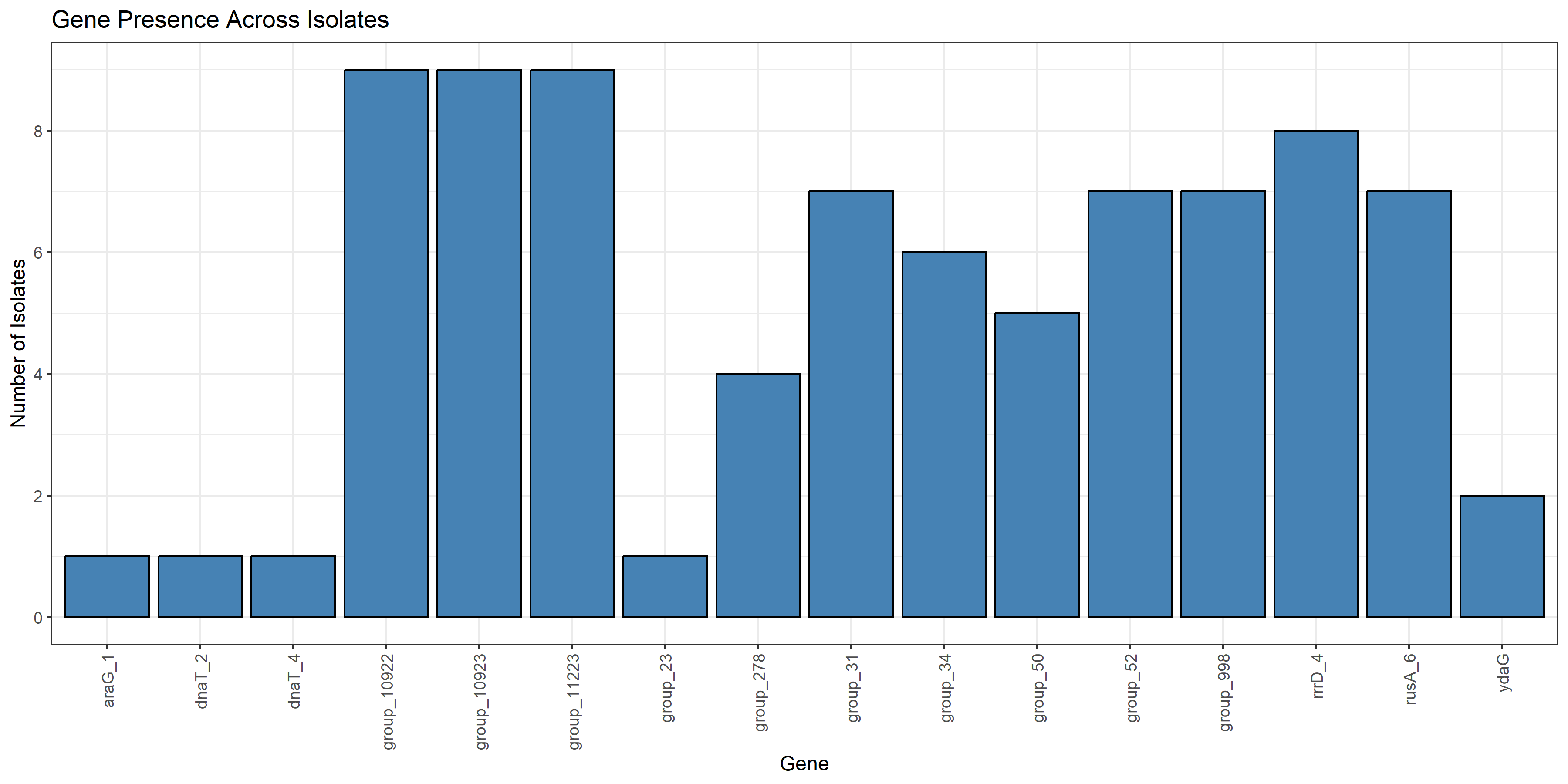
|
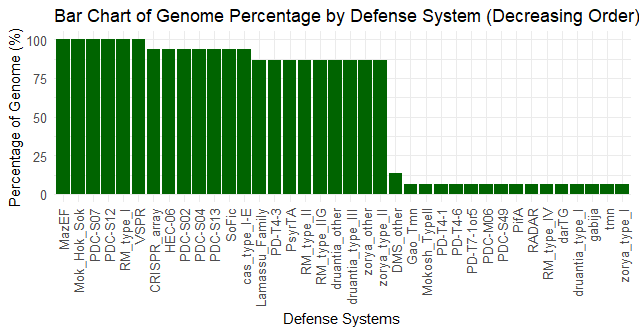
Fig 1. Barchart showing DS types vs percent genome |
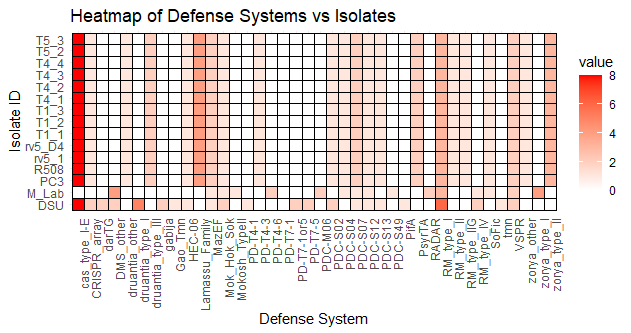
Fig 2. Heatmap showing type of DS in each phage resistant isolate |
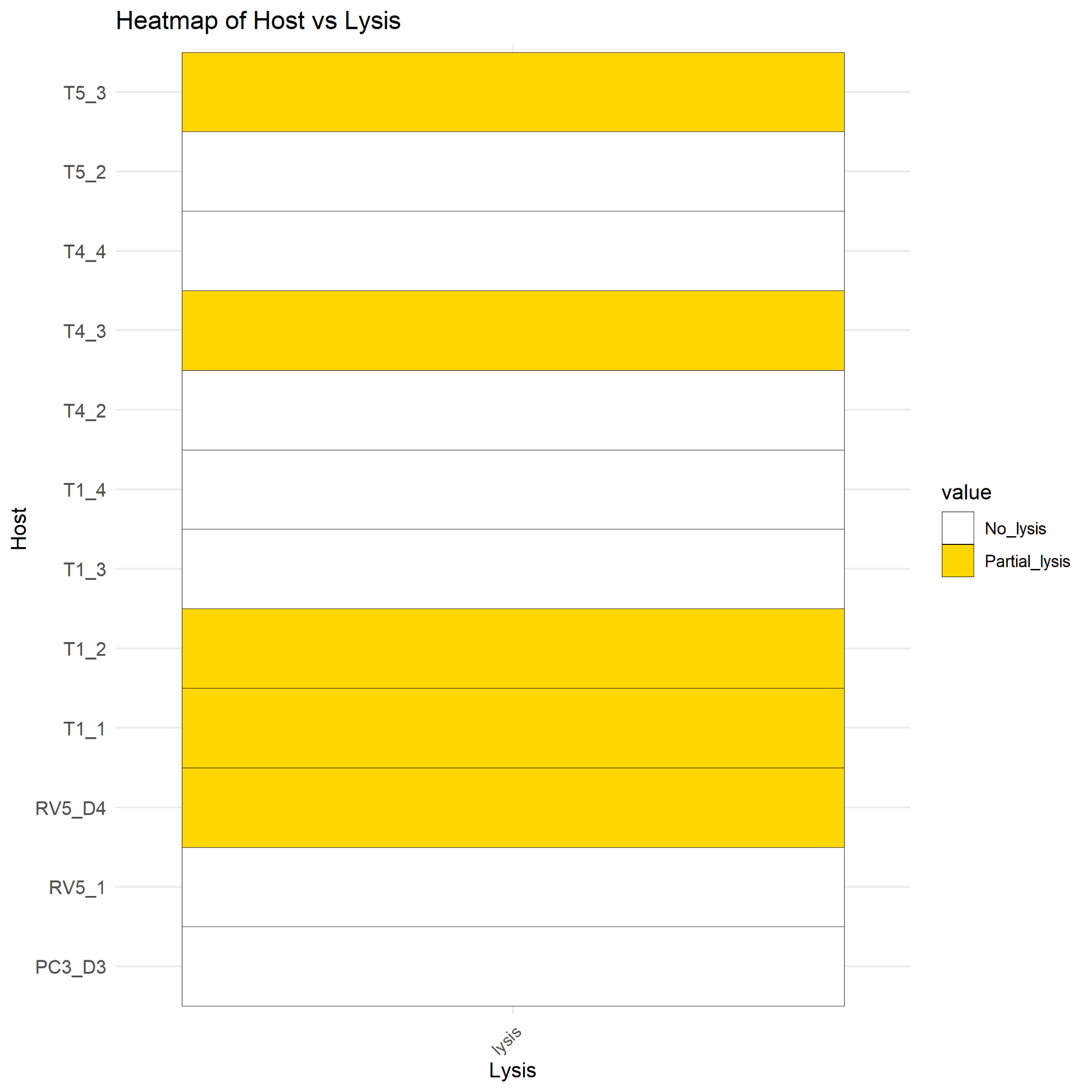
Fig 3. Heatmap of phage resistance and sensitivity against respective phages |
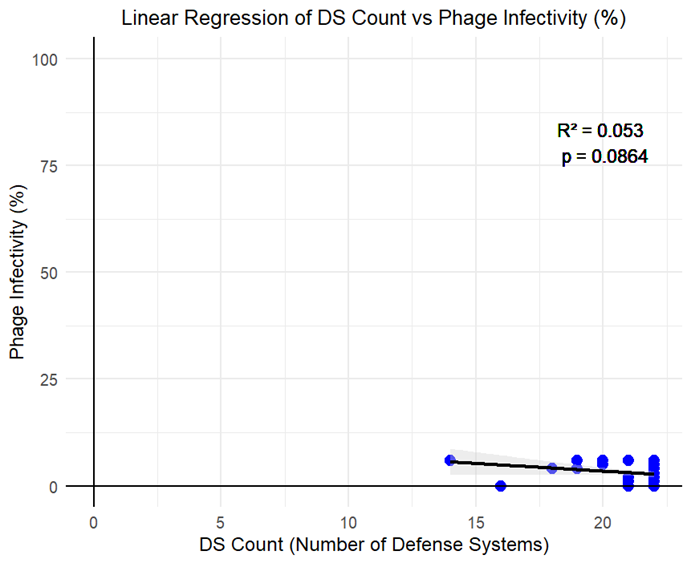
Fig 4. Relationship between phage infectivity and DS counts from phage mutant isolates |
Fig 5. Barchart showing gene loss in total no. of strains compared to reference strain R508 |
Fig. 6. Heatmap showing gene loss in each strain compared to reference strain |
Fig 7. Principal component analysis (PCA) on basis of gene presence absence showing similarities of mutants with reference strain
Figure 1 shows the percentage of isolates that contained each defense system within the genome. As there was a large discrepancy between the percentages, this shows that the isolates were relatively similar, which aligns with the fact that the isolates studied were E. coli O157 mutants collectively. Figure 2 shows how many of each defense system was present in each isolate, and the mutated isolates as well as reference strain R508 were all extremely similar. 5 out of 12 mutants isolated had partial lysis; T1_1, T1_2, T4_3, T5_3, and rv5_D4. These isolates show some formn of resistance development to the phages. Figure 4 shows a correlation between DS count and phage infectivity, although the data points are variable, so although there is a correlation, Figure 4 suggests an outside factor. Figure 5 shows the number of strains each gene was lost in. Figure 6 shows the isolates each gene was lost in, with blue indicating a loss between the isolate and reference strain R508. Figure 7 indicates that the mutants and reference strain R508 were still relatively similar in genetic composition.
Conclusion
Conclusions
- Gene absence may be attributed to resistance development of E. coli O157 infecting phages
- The phage resistance may be attributed to different defense systems observed in bacterial isolates, i.e., Restriction Modification system and CRISPR-Cas type IE
- The resistance can also be due to accessory gene loss and SNPs
Future Directions
- Current future directions would involve further research into multi-phage cocktails and combinations that would lead to greater resistance
- Study focused on single-phage impacts on isolates; multi-phage cocktails shown to be more effective in reducing phage resistance in isolates14
Citations
1. Hemolytic uremic syndrome (HUS). Mayo Clinic. Accessed November 1, 2024. https://www.mayoclinic.org/diseases-conditions/hemolytic-uremic-syndrome/symptoms-causes/syc-20352399
8. Kasman LM, Porter LD. Bacteriophages. In: StatPearls. StatPearls Publishing; 2024.
15. Wright RCT, Friman VP, Smith MCM, Brockhurst MA. Resistance evolution against phage combinations depends on the timing and order of exposure. MBio. 2019;10(5). doi:10.1128/mBio.01652-19
Acknowledgement
Dr. Dongyan Niu - Associate professor (University of Calgary)
Mawra Gohar - Ph.D. student
Jenny Hyun - Research associate
NSERC-Discovery Grant
Dr. Beatriz Garcia-Diaz (Webber Academy)