Hacking Evolution: Neutralizing Antibiotic Resistance with CRISPR
Grade 9
Presentation
Problem
The development of CRISPR technology has opened up a world of gene manipulation, bringing us closer to achieving the dream of achieving a world without sickness. However, with CRISPR still in its developmental stage and not ready for commercial use, antibiotic resistance continues to take millions of lives. Caused by misuse of prescribed drugs, antibiotic resistance is projected to kill 50 million people annually by 2050. To prevent the further progression of antibiotic resistance, scientists need to innovate and target the core of resistance; the genes. By integrating bio-circuits with CRISPR, not only will CRISPR research accelerate, but will make beating antibiotic resistance much more viable.
The contemporary state of CRISPR technologies exhibits extremely limited capabilities, being restricted to laboratory use, with several issues bewildering our most ingenious scientists. These issues include off-target effects, delivery challenges, cost, etc. This is why in my project I have proposed the use of bio-circuits. With bio-circuits possessing capabilities such as identifying resistant genes, signal amplification, etc, we can address concerns surrounding CRISPR use. While constructing a CRISPR-based bio-circuit requires multiple biological components and decades of research to be considered safe, my project revolves around the theoretical basic concept of building and testing a design of a CRISPR-based bio-circuit
Method
Refer to sections 6 through 6.4 in my research for the theoretical methodology details on how to build a CRISPR-based bio-circuit
Research
Antibiotic resistance is projected to kill 10's of millions of people in the years following up to 2030, becoming one of the leading causes of death worldwide. My research mainly focuses on how to beat antibiotic resistance by combining 2 revolutionary tools, CRISPR & bio-circuits. CRISPR-based bio-circuits allow for automation and effectiveness that's much more viable than manipulating CRISPR by hand. This method offers a 1-time solution to neutralizing resistance in a bacterial strain, almost eliminating the need for mass antibiotic research and production. The way my proposed circuit works is CRISPR is responsible for the actual gene editing (using Cas9 proteins to edit genes), while the bio circuit identifies which gene is causing resistance by identifying specific genetic markers in nucleotides. After the resistant gene has been found, the bio circuit activates CRISPR and "guides" it to the correct gene to make changes. My study is focused on the methodology of how to build the bio circuit and the information surrounding it.
2. How do Bacteria strains become resistant?
| Bacterial infection phases | |
|
2.1. Initial exposure |
When bacterial strains are initially introduced to antibiotics, the drug targets specific bacterial processes the organism needs to survive, such as inhibiting/preventing cell wall synthesis or protein production. Since it's the first time this bacteria has been exposed to the antibody, it works like a charm, inhibiting and killing the target bacteria effectively. However, because of the genetic variation in bacteria within a population, some bacteria may be able to survive and gain resistance. Because bacteria reproduce asexually, their daughter cell will be given their resistance to antibiotics as well. |
|
2.2. Acquiring Resistance |
There are multiple ways a bacterium may acquire resistance, my research covers the 3 most common ways they do this: Genetic Mutation - Because of environmental factors like radiation or chemicals, an error in DNA replication may happen. This could cause random new traits to appear, mutations like this may or may not cause more resistant genes to appear Binary Fission - This is the process in which bacteria reproduce asexually. When this process is in motion, bacterial DNA duplicates and divides into 2 genetically identical daughter cells. Even though this process had intentions to create clones, mutations during the replication process can begin to introduce genetic variation over time Transformation - It's a form of horizontal gene transfer where a bacteria picks up free DNA fragments from its environment (mostly from dead or lysed bacteria). If the foreign DNA is similar enough, homologous recombination will allow DNA to integrate into the bacterium's genome. Possibly giving the bacterium resistance or other survival mechanisms. Regardless of which way bacteria get resistance, conjugation (transfer of resistant gene via bacterial contact) & transduction (transfer of resistant genes via bacteriophages) allows bacteria to spread their resistant genes to other bacteria, making the drug once meant to kill bacteria useless |
|
2.3 Selection Pressure |
Selection pressure occurs when antibiotics are overused or misused (e.g., not completing the full treatment or using antibiotics for viral infections). It's basically when bacteria susceptible to antibiotics are killed off while the resistant bacteria survive. Impact? |
|
2.4. Spread of Resistance |
Bacteria aren't just idle, they transfer to humans the same way COVID did (human contact, environmental contamination, global travel, etc). As more and more people start to catch the infection, it makes the likelihood of catching the infection in an everyday setting higher, most likely causing slower recovery times and treatments that may not be effective. |
|
2.5. Multidrug resistance (MDR) |
As the name suggests, multidrug resistance (MDR) is when bacteria develop resistance against multiple antibiotics. After using multiple drugs to try killing an already resistant bacterial strain, there's a small chance it could gain immunity to other drugs as well. These bacteria often use a combination of different mechanisms to develop resistance, possibly creating its version of the same antibiotic, altering and changing its target. MDR is one of the biggest problems of doctors since they have limited options when it comes to curing this. In many cases, doctors use final-resort antibiotics, which are more expensive, less effective, and have stronger side effects. |
| 2.6. Potential Pandemic | In severe cases, a pandemic may begin if resistant bacteria can transfer from person to person quickly. In this stage, resistant bacteria will be in control of how fast it spreads, as without medication that works, there wouldn't be any way to progress the infection. |
3. What impact does Antibiotic Resistance have on the world?
3.1 Impact Statistics
- Around 10 million people are expected to die annually from antibiotic resistance by 2050
- With healthcare as poor as it is right now, Antibiotic Resistance will become a burden to countries all over the world
- Crops and agriculture suffer from new diseases constantly infecting crops
- People will start to doubt the healthcare system and become scared as incurable diseases take over the world

4. Why do current solutions not work?
- Vaccination
- Limitation of antibiotic use
4.3. Global Disparities:
5. CRISPR & Biocircuits
5.1. Useful terms
- 5.1.1 CRISPR - Gene editing tool used to precisely modify a piece of DNA
- 5.1.2 gRNA (guide RNA) - a short synthetic RNA molecule used in the CRISPR system-based genome editing
- 5.1.3 Cas9 - A protein derived from a bacterial immune system that's used as a tool to cut and modify DNA
- 5.1.4 Promoter - DNA sequence that begins the transcription of genes
- 5.1.5 Repressor - Protein that inhibits gene expression by binding to a promoter or operator region
- 5.1.6 Feedback loop - Amplifies a certain biological pathway or inhibits it, based on the outputs of a process that influence its own activity
5.2. What is CRISPR?
The CRISPR system primarily consists of:
5.2.1. Guild RNA (gRNA) - Responsible for guiding the Cas9 protein to the DNA within a genome that needs editing. By custom-designing RNA sequences that replicate the target DNA sequence, RNA can match with resistant DNA, signaling to Cas9 where to edit.
5.2.2. Cas9 Protein - It's an enzyme that acts as molecular scissors, editing/cutting DNA in the needed location. When the DNA is cut, natural cellular repair mechanisms can disable, replace, or correct faulty genes. This gives CRISPR the power to change basically anything within a living organism's genome.
5.3. What are Bio-Circuits?
- Sensing capabilities - They sense any threats within their environment by detecting genetic or environmental signals, such as antibiotic presence or the existence of resistant genes
- Response Activation - Once the threat is detected, they trigger responses to activate CRISPR to edit and disable target genes
- Control Systems - To program bio circuits and ensure they work, it can include regulatory mechanisms (e.g. Feedback loops, promoters, etc) to control the extent and timing of responses
Adding on to this some key components that ensure these functions are completed include:
- Sensors - Responsible for detecting molecular signals like antibiotic presence or gene expression levels
- Logic Gates - Processes information based on predefined conditions (e.g. activating CRISPR only when a threat is present)
- Actuators - Triggers responses within bacteria to combat antibiotic resistance (e.g. Gene editing, protein synthesis, cell death, etc)
Biocircuits, in my project, essentially act as a control panel for CRISPR. They can detect antibiotic-resistant bacteria and activate CRISPR when necessary. The biocircuit is a key system needed to increase the effectiveness of CRISPR.
5.4. How CRISPR-Based Bio-Circuits Work
CRISPR's modification system requires a specific sequence of steps to be followed in order to function. If these steps aren't done then CRISPR won't be able to edit genes precisely.
This is the following process:
- Gene Identification - Firstly, to ensure CRISPR edits the right gene, we need to know which gene is responsible for the resistance. This could be done by using gRNA, as its job is to essentially scan for resistant sequences within bacteria genes. Once the target sequence is found we can edit it.
- DNA Binding - Now the nucleic sequence has been found, the gRNA will begin to bind to the complementary DNA, forming a stable complex.
- Cas9 Editing - Because of the gRNA binding to the DNA, Cas9 will know where to edit the gene without making mistakes. Cas9 can stop resistance by either cutting, replacing, or binding to the DNA.
- Cellular Dysfunction - If the DNA was cut then the bacterial cell senses something wrong, and tries to fix the break. This ends up disrupting the resistant gene or completely removing the harmful genes entirely. However, sometimes but very rarely, a bacteria with a strong repair system could potentially repair its DNA to be resistant again. On the other hand, if Cas9 binds to the DNA, the cell will sense nothing wrong and will simply lose its resistance as RNA polymerase won't be able to read the gene, preventing it from being expressed.
- Cellular Response - Now once the antibiotics are reintroduced into a person's body, these bacteria will no longer have resistance.
6. Hypothesis
If we're able to apply CRISPR-based bio-circuits to resistant bacteria then we'll be able to neutralize antibiotic resistance because by using synthetic bio circuits we can precisely target and disable resistant genes that are responsible for antibiotic resistance
6.1. Variables
Manipulated - Presence of antibiotics (Ampicillin or tetracycline or E.coli)
Responding - CRISPR activity (wether or not antibiotic genes are edited)
Controlled - Only E.coli strain, amount of antibiotic, experiment conditions, plasmid
7. Creating a Theoretical CRISPR Based Bio-Circuit - (Methodology)

- Sensors
- Create a genetic sensor capable of detecting blaTEM gene sequences within E.coli
- Add gRNA to recognize, then bind to blaTEM
- The sensor should be tested to make sure it can differentiate between resistant and non-resistant E.coli strains
- Promoter
- Have the promoter activate only in the presence of antibiotic-resistant E.coli
- Must transcribe the CRISPR-cas9 system reasonably to the quantity of blaTEM genes detected
- Feedback loops - Regualtes CRISPR's responses
- Positive feedback loops ensure CRISPR stays activated until the resistant gene is eliminated
- Negative feedback loops add safety; preventing unnecessary gene editing when blaTEM is gone
- Repressors - Controls activation
- Act as a fail-safe mechanism, inhibiting CRISPR activation when resistance isn't detected
- Basically makes sure the right genes are modified
7.3. Step 3: Implement The Circuit
After we've made sure the bio circuit can identify blaTEM, it must neutralize resistance by activating CRISPR. For it to work properly, we need to make sure it can:
Correctly Identify genes - The gRNA will only bind to the resistant gene within E.coli & not anything else
Bind to DNA & Activate Cas9 - gRNA should be able to guild Cas9 to the resistant gene site
Deactivate/Disrupt Genes - Cas9 should be able to edit blaTEM and render it useless
Have a Positive Outcome - Make sure that the E.coli is once again resistant to antibiotics
7.4. Step 4: Test & Modify
Once experiments have been conducted and the circuit has been created, we need to test it thousands of times to make sure it works & is safe:
- In-Vitro Testing
- After placing our CRISPR-based bio-circuit into E.coli cultures, observe gene editing efficiency using PCR and sequencing techniques
- Test to confirm that E.coli has lost its antibiotic resistance after treatment
- Check For Mutations
- Analyze the bacteria's genome to ensure potential mutations or other unintended effects are avoided
- Make sure it is only blaTEM that's being edited
- Add Finishing Touches
- Adjust the mechanics of the circuit if deemed necessary (e.g promoter sensitivity, modify feedback loops, etc)
8. Pros & Cons of Antibiotic Resistance
Antibiotic resistance is a problem that concerns not only us but the entire world. However, with CRISPR on the rise, it is expected to neutralize antibiotic resistance completely. But like all good things, there are both positive and negative sides to this gene editing tool. On the positive side, since these bio-circuits remove antibiotic resistance from bacteria, they remove the need for operations to continue creating new antibiotics, which is costly and time-consuming. Furthermore, their automation provides real-time bacterial detection, making treatment much more effective. Not to mention that CRISPR-based bio-circuits can be adapted to the resistant genes of multiple bacterial strains, allowing for a long-term, targeted, versatile solution.
However, there are further challenges that need to be addressed. For example, delivering the CRISPR circuit to the resistant bacteria remains a technical problem we still struggle with, as bacteria might have the ability to evade genetic editing. Some scientists have concerns about target editing, which could potentially lead to unprecedented consequences. Finally, to be able to implement CRISPR on a large scale, we still need extensive testing & research, requiring a large sum of money (mostly from funds), making CRISPR a technology that most likely won't be implemented within the near future. Just as space exploration leads to technological advancements, researching CRISPR will unveil new technologies & innovations that will positively impact health for generations to come
Data
Dataset:
Figure 2. The bar graph represents deaths caused by antibiotic resistance
1.2 million deaths are caused each year by Antimicrobial resistance, the majority of which are antibiotic-resistant deaths
Figure 2.1. Line graph representing the chance of bacteria developing resistance
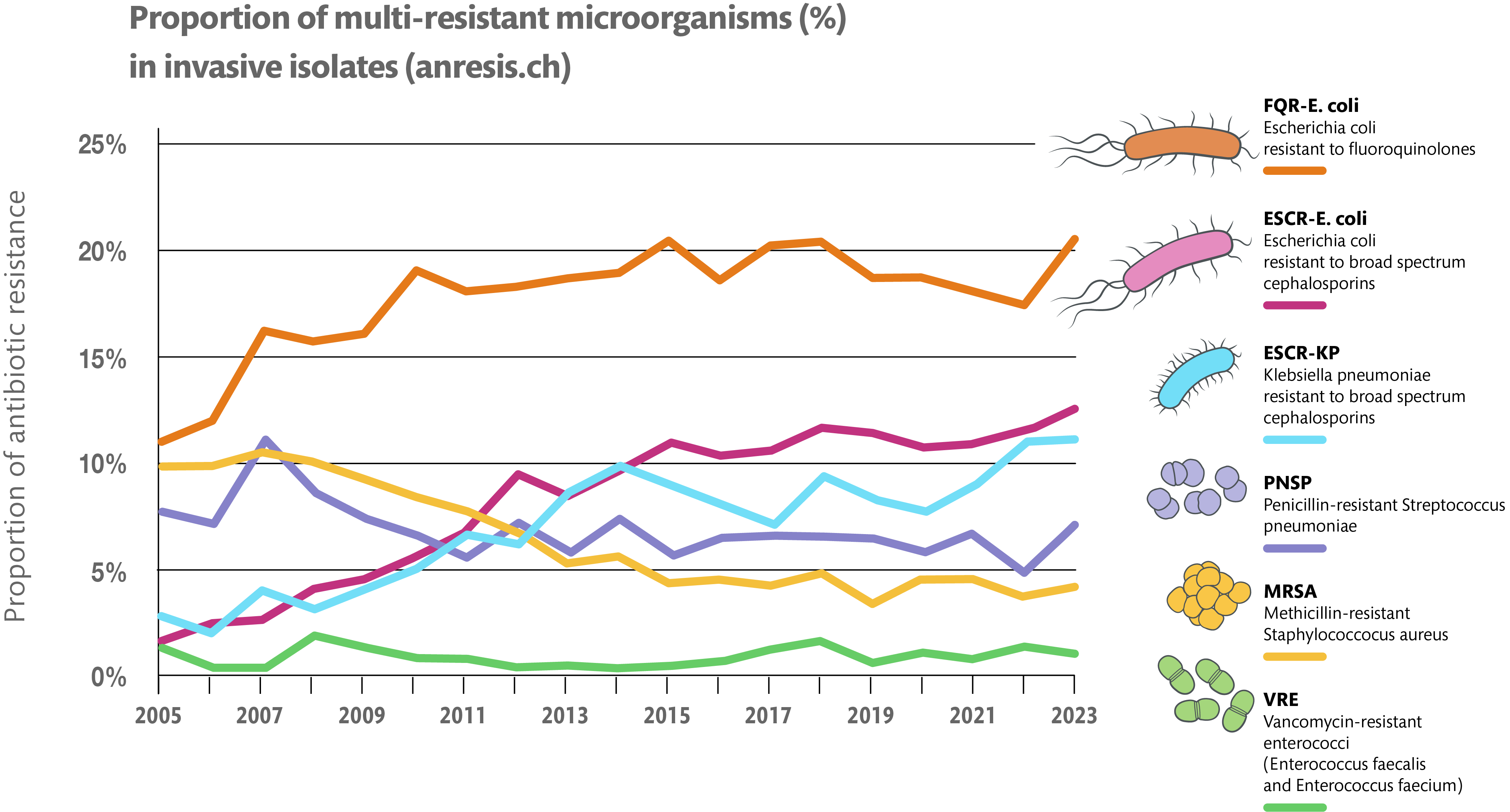
Over the past 20 years, E.coli has had an increase in resistance by more than 10%. By 2050, this number along with other bacteria resistance, is expected to increase
Conclusion
Conclusion
In conclusion, if we’re able to fully develop CRISPR-based bio-circuits to accurately and efficiently identify and edit resistant genes, we can neutralize antibiotic resistance in its entirety. Introducing new technologies and finally replacing our outdated method of simply creating more antibiotics to cure one version of a bacterial strain of the countless other bacterial strains out there. The intended end result of my research was to create a theoretical CRISPR-based bio-circuit that could edit genes in bacteria without struggle. While this was all theoretical, my project gives a simple yet detailed framework on what's needed to build the circuit and how to test it. However, making a bio-circuit in real life will require a lot more research and testing before being implemented in hospitals.
In the future, I hope my project will inspire even just one person to become involved in the medical field to explore CRISPR and bio-circuits. Because the potential is astronomically huge, as the ability to edit genes won’t only neutralize antibiotic resistance, but will inspire breakthroughs in other areas of medicine too.
Ultimately, with the possibilities of CRISPR being endless, it's just a matter of our imagination and how much we’re willing to innovate. With antibiotic resistance continuously becoming a bigger and bigger problem that needs to be addressed, I hope people will look to the future and try to contribute by funding or becoming a researcher of CRISPR.
Citations
Scholarly Articles
- Nouri, Ahmad, et al. “CRISPR-Based Platforms for Antibiotic Resistance: Detection and Therapeutic Approaches.” Frontiers in Bioengineering and Biotechnology, vol. 10, 2022, https://www.frontiersin.org/articles/10.3389/fbioe.2022.869206/full.
- Zhang, Wei, et al. “Antibiotic Resistance and Its Mechanisms: New Insights into an Old Problem.” Scientific Reports, vol. 11, 2021, https://www.nature.com/articles/s41598-021-96735-4.
- Yan, Ming, et al. “A Novel dCas9-Based Gene Regulation System with Enhanced Targeting Efficiency.” Nature Communications, vol. 13, 2022, https://www.nature.com/articles/d41586-022-00228-x.
- “Dead Cas9 (dCas9): Concept, Functions, and Applications.” Genetic Education, https://pmc.ncbi.nlm.nih.gov/articles/PMC9356603/.
- “Antibiotic Resistance Mechanisms: Understanding the Evolution of Bacterial Defenses.” PubMed Central, https://pmc.ncbi.nlm.nih.gov/articles/PMC8549092/.
- “CRISPR and Bio-Circuitry: A New Era of Genetic Engineering.” PubMed Central, https://pmc.ncbi.nlm.nih.gov/articles/PMC6152938/.
- “Genetic Circuit-Based CRISPR Systems for Targeted Gene Editing.” PubMed Central, https://pmc.ncbi.nlm.nih.gov/articles/PMC1091604/.
- “Gene-Targeting Strategies for Antimicrobial Resistance.” PubMed Central, https://pmc.ncbi.nlm.nih.gov/articles/PMC10573330/.
Government and Health Organization Reports
- Centers for Disease Control and Prevention. “About Antimicrobial Resistance.” CDC, https://www.cdc.gov/antimicrobial-resistance/about/.
- Centers for Disease Control and Prevention. “Causes of Antimicrobial Resistance.” CDC, https://www.cdc.gov/antimicrobial-resistance/causes/index.html.
- Government of Canada. “About Antimicrobial Resistance.” Public Health Agency of Canada, https://www.canada.ca/en/public-health/services/antimicrobial-resistance/about.html.
- World Health Organization. “Antimicrobial Resistance.” WHO, https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
News Articles and Reports
- “Stanford Explainer: CRISPR Gene Editing and Beyond.” Stanford News, 2024, https://news.stanford.edu/stories/2024/06/stanford-explainer-crispr-gene-editing-and-beyond#how-CRISPR.
- “More than 3.9 Million Deaths from Antibiotic-Resistant Infections.” Institute for Health Metrics and Evaluation (IHME), https://www.healthdata.org/news-events/newsroom/news-releases/lancet-more-39-million-deaths-antibiotic-resistant-infections.
- “Antimicrobial Resistance (AMR).” The Lancet, vol. 403, no. 10429, 2024, https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(24)01867-1/fulltext.
Educational and Research Resources
- “Promoter.” National Human Genome Research Institute, https://www.genome.gov/genetics-glossary/Promoter.
- “Plasmid.” National Human Genome Research Institute, https://www.genome.gov/genetics-glossary/Plasmid.
- “Targeting Antibiotic-Resistant Microbial Infections with Synthetic Biology.” Integrated DNA Technologies, https://www.idtdna.com/pages/education/decoded/article/targeting-antibiotic-resistant-microbial-infections-with-synthetic-biology.
Multimedia and Video Sources
- YouTube. “CRISPR Gene Editing Explained.” YouTube, uploaded by Kurzgesagt – In a Nutshell, https://youtu.be/ZvhFeGEDFC8.
- “Medicine Pills and Tablet Macro View.” Stock Video, https://dm0qx8t0i9gc9.cloudfront.net/thumbnails/video/Vd3bj2jPe/videoblocks-medicine-pills-and-tablet-macro-view-of-ph.
Additional Scientific Database Sources
- “Antibiotic Resistance Trends.” PNAS, 2025, https://www.pnas.org/doi/10.1073/pnas.2411919121.
- “Mechanisms of Antibiotic Resistance.” Molecular Cancer, https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-023-01925-5.
- “Gene Editing for AMR Treatment.” Nature Communications, https://www.nature.com/articles/s41467-021-21740-0.
- Microbe Notes. E. coli on Blood Agar., 2017, https://microbenotes.com/wp-content/uploads/2017/07/E.-coli-on-Blood-Agar-768x614.jpg.
- Beckman Coulter Diagnostics. "Antibiotic Resistance Infographic." Beckman Coulter Diagnostics Blog, 2025, https://media.beckmancoulter.com/-/media/diagnostics/blog/images/diagnostics/micro-amr-blog/antibiotic-resistance-inforgrahic-1200x900.jpg?rev=a2823a158ea34a3e9cc79816df979bbb&hash=003CFBFA46334A27CBD51F7267458BDC.
- Thanks to SlidesGo for my template
- Chat GPT
For my project I utilized generative AI tools to help enhance my project in various ways. While every foundational idea, research, and insight was a product of my hard work, AI helped structure and organize my work in a more absorbable fashion, and also wrote links in MLA format. I have ensured all info collected is from reliable sources and my trifold presentation hopes to reflect my understanding of my topic. AI is revolutionary and is a tool that must used appropriately, and that is exactly how I used it. All sources I have used have now been properly sourced, including any AI I may have used.
Acknowledgement
Thank you to
My friend Ranbir for helping me think of what I was passionate in, leading me to discover this project
My teacher Ms.Shoults for answering any questions I may have had throughout this whole confusing process
my parents for supporting me the whole way, letting me skip lessons to work on my project
And finally my buddy Mirgank for always pushing me to strive harder toward my goals, even when I started struggling and seemed hopeless (the last one to submit)

