Shining Light on E. Coli: Investigating the Effects of UV-C Radiation on Bacterial Growth
Aarav Pardeshi Yatharth Trivedi
Grade 8
Presentation
Hypothesis
If we expose K12 Escherichia coli to UV-C radiation for different durations at a distance of 20 centimeters, then the minimum amount of time to fully inactivate K12 E. coli cultivation will be 60 seconds because at this duration, the nucleotide excision repair mechanism would be unable to keep up with the recurring cyclobutane pyrimidine dimers and 6-4 photoproducts, leading to the absence of binary fission due to cell death.
Research
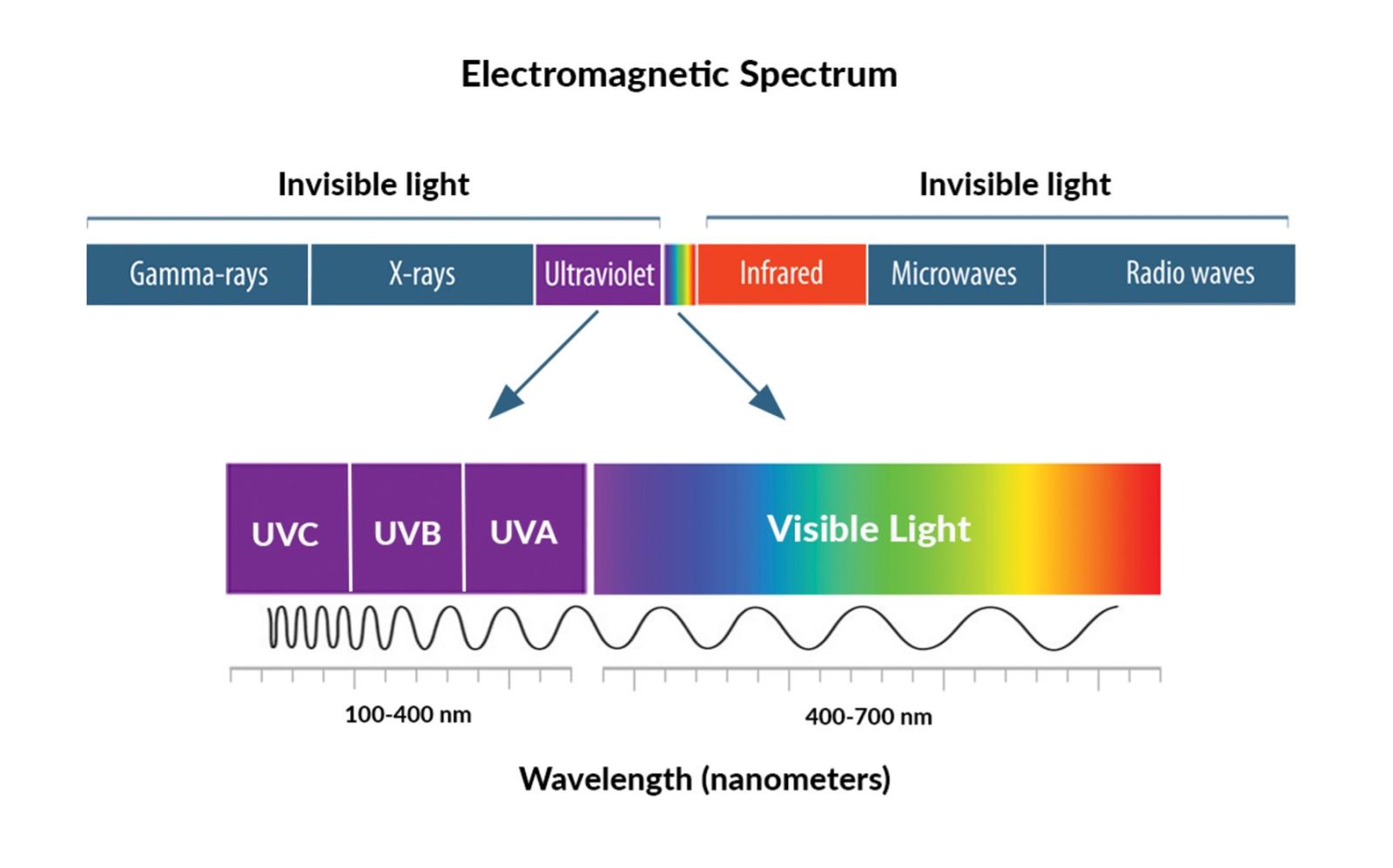
Ultraviolet Light
Ultraviolet (UV) light is a source of electromagnetic radiation consisting of wavelengths between 10 and 400 nm. Therefore, these wavelengths are longer than X-rays but shorter than visible light. Roughly 10% of the sun’s light emissions consist of UV radiation. It is often generated by specialized light which includes mercury-vapor lamps, germicidal lamps, black lights, etc. Long-wavelength forms of UV light like UV-A and UV-B are not forms of ionizing radiation (separation of electrons from respective atoms) due to insufficient energy carried by their photons. However, the shortest wavelengths of UV-C light which are the most energetic, can be considered as ionizing radiation.
When radioactive material emits UV radiation, it is categorized into 3 main forms,
- UV-A: has the longest wavelengths (320-400 nm)
-UV-B: has medium wavelengths (280-320 nm)
-UV-C: has the shortest wavelengths (100-280 nm)
The word “ultraviolet” originates from Latin where “ultra” means “beyond”. Therefore ultraviolet translates into “beyond violet”. That is because ultraviolet is even more energetic (shorter wavelengths) compared to violet. UV radiation was founded by a German physicist Johann Wilhelm Ritter in 1801. He referred to this discovery as “oxidizing rays” to differentiate it from “heat rays”. “Oxidizing rays” were later modified to “chemical rays” during the 19th century. However, some scientists believed that this was a completely different type of radiation and should not be considered light. Therefore, John William Draper called these “tithonic rays”. Later on, the name chemical rays was changed to ultraviolet, and heat rays were known as infrared radiation. The effectiveness of highly energetic light on microorganisms such as bacteria was determined in 1878. The impact of UV radiation on DNA was discovered in 1960. In 1893, German physicist Victor Schumann discovered “vacuum ultraviolet”. This was UV radiation under 200 nm which was actively taken in by the air.
Protecting your eyes from the sun’s UV light. (2022, July 5). National Eye Institute. Retrieved January 6, 2024, from https://www.nei.nih.gov/about/news-and-events/news/protecting-your-eyes-suns-uv-light
Deoxyribonucleic and Ribonucleic Acids
Deoxyribonucleic acid (DNA): an extensive macromolecule that carries an organism's genetic information. “Deoxy” refers to the absence of a hydroxyl group in the ribose sugar ring of each nucleotide in DNA.
Ribonucleic acid (RNA): transcribed deoxyribonucleic acid
Nucleic acids are extensive polymers composed of contiguous nucleotides. Nucleotides are kind of like the building blocks of DNA and RNA. They consist of ribose sugar and one phosphate group forming the backbone, along with one purine (adenine or guanine) or pyrimidine (thymine or cytosine) base, creating base pairs. In DNA, the bonding of bases between opposite strands can only be as follows, thymine-adenine, and cytosine-guanine. However, in RNA, all thymine bases of a gene are substituted by uracil.
Next, phosphate-sugar linkages forming the backbone are connected by phosphodiester bonds. This happens when the 3’-hydroxyl group of a specific sugar is connected to the 5’-hydroxyl group of the sugar in the adjoining nucleotide. These linkages are important because they determine the direction in which a polynucleotide (repeating nucleotides) should be interpreted. These two nucleic acids have different structures, as DNA is considered a double helix (two strands or two polynucleotide chains twisted around each other) and RNA is single-stranded.
Latham, K. (2021, January 9). DNA vs. RNA. Biology Dictionary. Retrieved November 15, 2023, from https://biologydictionary.net/dna-vs-rna/
Escherichia coli
Escherichia coli (E. coli) is an example of bacteria found in the lower intestines of animals and humans. It is also sometimes detected in food products. However, certain subtypes of E. coli like K-12 and B are considered nontoxic. E. coli can be grown in colonies which are observable concentrations of microbial cells developing from a single cell. This microorganism is considered a prokaryote.
Prokaryotes are single-celled organisms that lack a nuclear membrane (still have some nuclear components) and membrane-bound organelles. Organisms such as bacteria and archaea are the two types of prokaryotes.
Prokaryotic cells reproduce through a type of cell division called binary fission. This happens when the parent cell is divided into two daughter cells. In this process, DNA replicates, to form a new copy. Both of these DNAs then move to opposite ends of the cell. Next, as the cell begins to enlarge, a septum which is a cell wall forms in the middle dividing the parent cell into two daughter cells. This process is then repeated to make bacteria grow.
Nutrition of Bacteria - Structure of Bacteria. (2021, September 22). DesktopClass.com. Retrieved January 6, 2024, from https://www.desktopclass.com/biology/structure-of-bacteria-part-4-f-sc-biology-chapter-6/
DNA Replication and Gene Expression
-
Gene Expression
Gene expression is the process that cells undergo to become translated into amino acids which are required for proteins. In prokaryotes, this occurs in the cytoplasm through two major sequences, transcription and translation.
The first step to transcription is DNA unwinding in the transcription bubble. Then, enzymes called RNA polymerase connect to a promoter in a template strand which is a single strand of DNA consisting of the genes being transcribed. A promoter is the starting sequence of bases in a specific gene. Next, RNA polymerase interprets the template strand from 3’ to 5’, resulting in the creation of an mRNA (messenger RNA) molecule. An mRNA molecule is the same as the non-template strand of DNA, except the thymine bases are replaced with uracil bases. Finally, the last step in this process is termination. In prokaryotes, termination can either be rho-independent or rho-dependent. In rho-independent termination, the mRNA strand naturally detaches from the template DNA strand. However, in rho-dependent termination, the rho protein attaches to the mRNA molecule and separates it from the DNA template strand.
The second step is translation. mRNA molecules are used to form amino acids which are the building blocks of protein. Three consecutive bases of a mRNA molecule (codon) are what tell the ribosome which amino acid shall be generated next. A ribosome is like a machine found in the cytoplasm of prokaryotes used to convert mRNA into proteins. Next, transfer RNA (tRNA) brings amino acids to the ribosome. Several amino acids linked together are called a polypeptide chain which then form proteins. Finally, a stop codon terminates the translation process as it does not have a complementary molecule of tRNA because it does not code for any amino acid.
-
DNA Replication
DNA replication can be divided into three parts.
-
The double helix structure is unzipped by an enzyme called DNA helicase. This is caused by helicase separating base pairs via the weakening of hydrogen bonds.
-
The two polynucleotides from the unzipped double helix act as a template for the formation of a new complementary polynucleotide. This process begins with a primer, which is a small RNA polynucleotide attaching to the 3’ end of an original DNA strand. Next, an enzyme called DNA polymerase III adds nucleotides to the primer complementary to the original DNA strand going from 5’ to 3’. This newly synthesized DNA strand is called the leading strand. The complementary DNA strand to the other original polynucleotide is called the lagging strand since it is generated in fragments. This is because the DNA polymerase can only interpret DNA sequences in the 5’ to 3’ direction. As these strands are antiparallel, the lagging strand moves opposite to the leading one. Once both strands have been synthesized, an enzyme called ligase joins the last fragments as synthesizing them would result in DNA polymerase going in the wrong direction.
-
The leading and lagging strands coil around their respective polynucleotide to form a double helix. This form of replication is known as semi-conservative since every DNA molecule would consist of one old polynucleotide/DNA strand.
UV-C Effects on DNA
-
Cyclobutane pyrimidine dimers (CPD): when two vicinal pyrimidine bases on a polynucleotide adjoin by a cyclobutane ring. A cyclobutane is a simple ring composed of four conjoining carbon atoms.
-
6-4 photoproduct (6-4 PP): when two adjoining nitrogenous bases of DNA form a covalent bond between the carbon atom at the sixth position of one base and the carbon atom at the fourth position of the adjacent base.
-
Dimers can prevent RNA and DNA polymerase from interpreting the genetic code correctly. Thus, processes of replication and gene expression are misled potentially leading to the death of the cell.
Pyrimidine dimer. (n.d.). Wikipedia. Retrieved November 15, 2023, from https://en.wikipedia.org/wiki/Pyrimidine_dimer
Nucleotide Excision Repair
Nucleotide excision repair (NER) is a repair mechanism that can fix DNA damage generated by ultraviolet radiation like CPDs or 6-4 PPs. In Escherichia coli, this process is operated by a network of proteins and enzymes called the uvrABC endonuclease. This mechanism involves the proteins, UvrA, UvrB, and UvrC along with the enzyme DNA helicase II. The first step in NER is when UvrA and UvrB proteins analyze the polynucleotides while UvrA detects any distortions in the double helix structure. When a pyrimidine dimer is identified the UvrA protein leaves and the UvrC protein arrives to connect with the UvrB. These proteins cleave 12 nucleotides as UvrB cuts the phosphodiester bond of 4 nucleotides directed towards the 3’ end from the DNA damage, and UvrC cuts the phosphodiester bonds of 8 nucleotides directed towards the 5’ end from the DNA damage. However, to fully remove this segment of 12 nucleotides, DNA helicase II eliminates the hydrogen bonds between the base pairs. Finally, the consequent space from the excised 12 nucleotides is filled by DNA polymerase, and ligase seals both ends of this new strand with the polynucleotide.
However, nucleotide excision repair not being able to keep up with excessive DNA damage can result in the death of the bacterial cell because of incorrect DNA replication. Additionally, unrepaired DNA damage leads to errors in the transcription process as the DNA sequence of genes cannot be interpreted by RNA polymerase due to pyrimidine dimers.
Variables
Independent:
- Duration of UV-C light exposure on K12 Escherichia coli samples
Dependent:
- Growth of E. coli colonies (we will measure this by counting the amount of colonies per mm² using a laminated millimeter grid paper)
Controlled:
- Distance of nutrient agar plates from UV-C source
- Wavelength of UV-C light
- Time spent in enclosed area
- Temperature of enclosed area
- Strain of E. coli
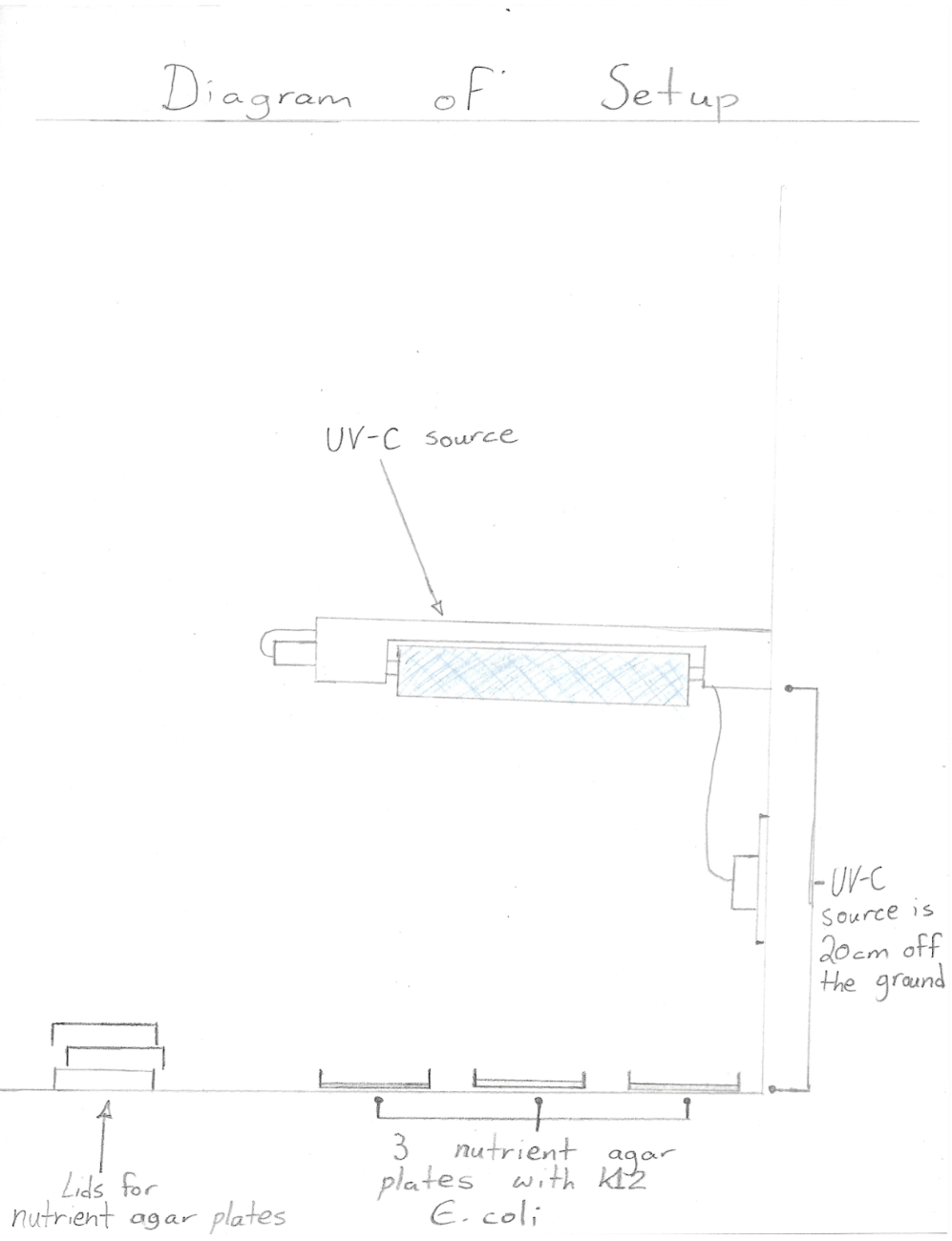
Procedure
- Wear nitrile gloves on both hands and put on safety goggles
- Swab K12 Escherichia coli on the designated side of all 18 nutrient agar plates with sterile cotton swabs in a consistent, zigzag pattern
- Place 3 nutrient agar plates labeled as “control” in enclosed area to serve as a control for the experiment
- Place 3 nutrient agar plates labeled “15” 20 cm below the UV-C source
- Turn on the UV-C source for 15 seconds and then switch off (relocate the UV-C treated agar plate to enclosed area)
- Place 3 nutrient agar plates labeled “30” 20 cm below the UV-C source
- Turn on the UV-C source for 30 seconds and then switch off (relocate the UV-C treated agar plate to enclosed area)
- Place 3 nutrient agar plates labeled “60” 20 cm below the UV-C source
- Turn on the UV-C source for 60 seconds and then switch off (relocate the UV-C treated agar plate to enclosed area)
- Place 3 nutrient agar plates labeled “120” 20 cm below the UV-C source
- Turn on the UV-C source for 120 seconds and then switch off (relocate the UV-C treated agar plate to enclosed area)
- Place 3 nutrient agar plates labeled “300” 20 cm below the UV-C source
- Turn on the UV-C source for 300 seconds and then switch off (relocate the UV-C treated agar plate to enclosed area)
- Leave all 18 agar plates in enclosed area at approximately 22 degrees Celsius for 48 hours
- After 48 hours, observe K12 E. coli growth by measuring the total area covered by colonies on each nutrient agar plate. (Use the laminated millimeter grid by placing it on the bottom of each nutrient agar plate to record the size of K12 Escherichia coli growth in mm²)
- Place all equipment that has come into contact with Escherichia coli into a labeled medical waste bag
- Dispose of all biohazardous materials in a commercial autoclave
Observations
RESULTS:
Total Area Covered by K12 E. coli Colonies when Exposed to Different Durations of UV-C
|
Duration of UV-C Exposure (seconds) |
Total Covered Area by Colonies (cm²) (Agar Plate 1) |
Total Covered Area by Colonies (cm²) (Agar Plate 2) |
Total Covered Area by Colonies (cm²) (Agar Plate 3) |
Average Area Covered by Colonies (cm²) |
Inactivated K12 E. coli growth (%) |
|
0 (control) |
51.5 |
49.5 |
50 |
50.33 |
0 |
|
15 |
46.5 |
50 |
47 |
47.83 |
4.97 |
|
30 |
30 |
28.5 |
30.5 |
29.67 |
41.05 |
|
60 |
1 |
1.5 |
14.5 |
8.5 |
83.11 |
|
120 |
0 |
0.5 |
0.5 |
0.33 |
99.34 |
|
300 |
0 |
0 |
0 |
0 |
100 |
Inactivation Percentage: This is the amount of inactivated K12 E. coli growth, meaning how much bacterial growth was reduced in terms of percentage when comparing average colony growth in control plates to a specific duration. This is calculated by taking the average area covered by colonies of a specific duration and dividing it by the average area covered by colonies in control plates (50.33 cm²). Next, the quotient is multiplied by 100, and then, subtract 100 from the product. Lastly, the final figure is rounded to the nearest hundredth if possible.
Qualitative Observations:

CONTROL
- Absence of individual colonies
- K12 E.coli growth appeared as per the streaking pattern
- Presence of bubbles within nutrient agar in 3 plates
- Streaks appear in a white color to differentiate from nutrient agar
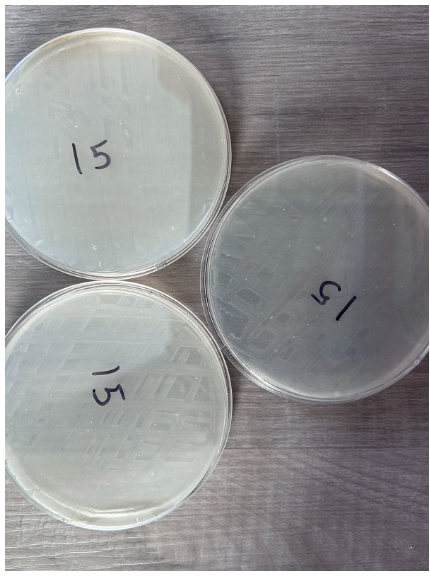
15 Seconds
- Absence of individual colonies
- K12 E.coli growth appeared as per the streaking pattern
- Presence of bubbles within nutrient agar in 3 plates
- Streaks appear in a white color to differentiate from nutrient agar
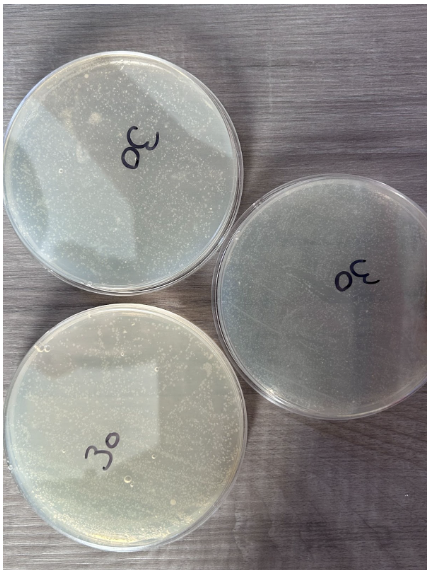
30 Seconds
- Presence of individual colonies only
- Scattered K12 E. coli colony growth that does not follow the streaking pattern
- Presence of bubbles within nutrient agar in 2 plates
- Colonies appear in a white color to differentiate from nutrient agar
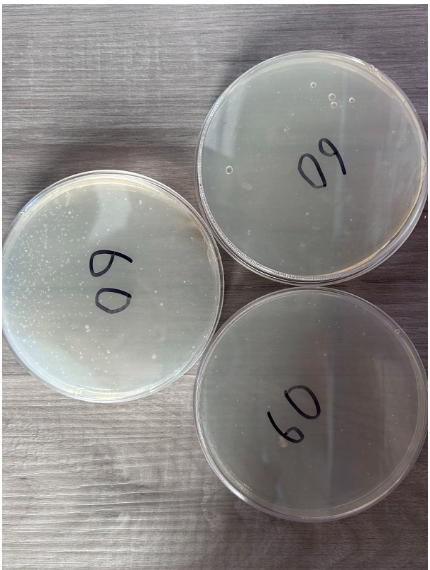
60 Seconds
- Presence of individual colonies only
- Scattered K12 E. coli growth occurred with colonies spread apart in two nutrient agar plates
- Presence of bubbles within nutrient agar in 1 plate
- One nutrient agar plate contains a high concentration of colony growth in one half
- The other two nutrient agar plates have little to no colony growth
- Colonies appear in a white color to differentiate from nutrient agar
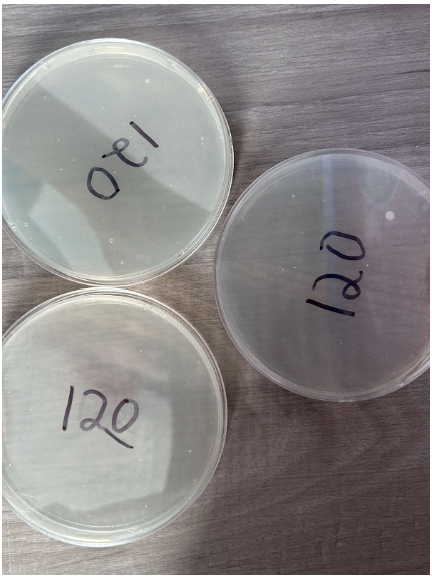
120 Seconds
- Minimal presence of individual colonies
- One of the nutrient agar plates consists of a fairly large colony measuring 0.13 cm²
- Low concentration of colonies within a given area
- Colonies appear in a white color to differentiate from nutrient agar
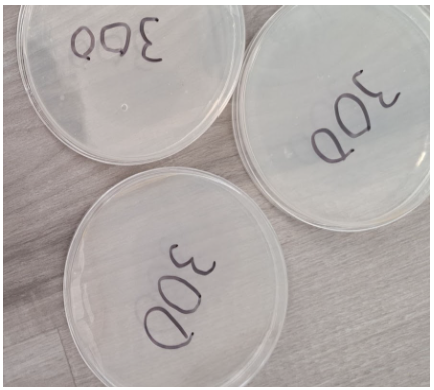
300 Seconds
- Absence of individual colonies
- Presence of a bubble within the nutrient agar in 1 plate
Analysis
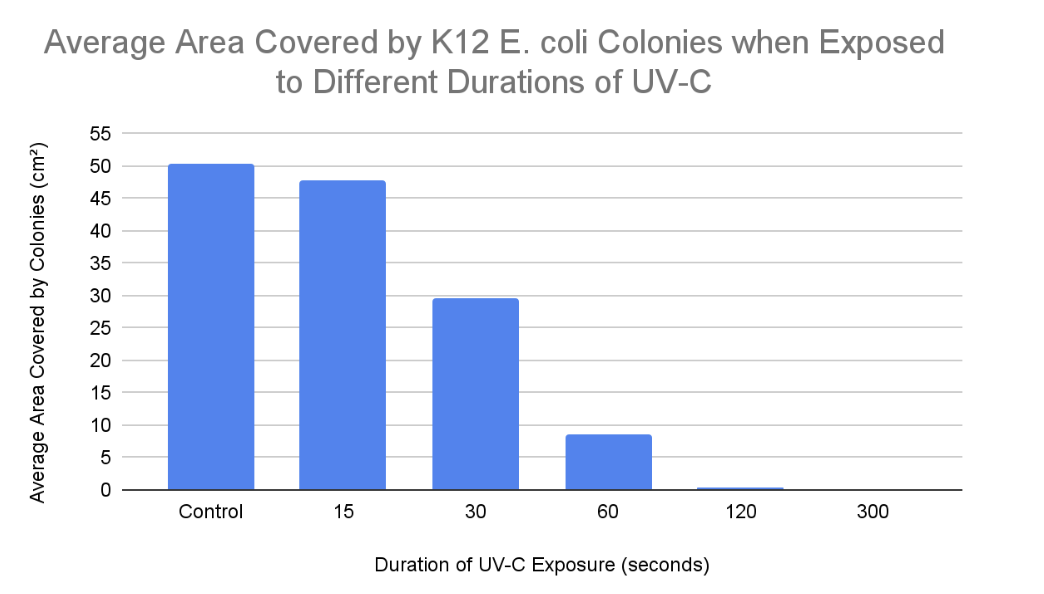
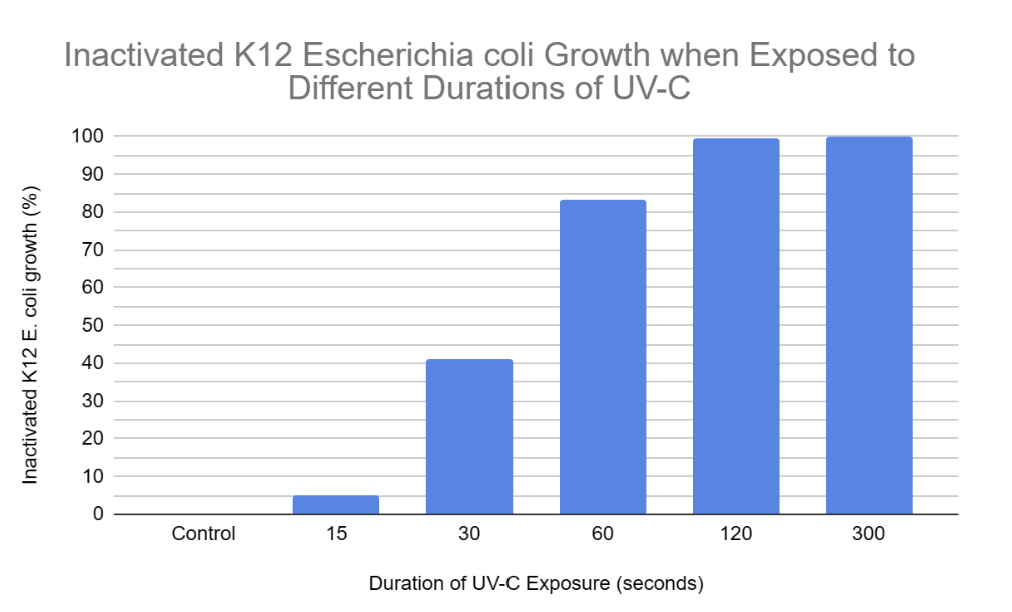
According to the results the trend was as expected, because as K12 E. coli was exposed to UV-C for increased amounts of time, the amount of colony growth decreased. In other terms, the percentage of inactivated K12 E. coli growth increased as it was exposed to higher durations of UV-C. This is because longer UV-C exposure resulted in more frequent cyclobutane pyrimidine dimers and 6-4 photoproducts resulting in the nucleotide excision repair mechanism being unable to keep up with the damage. Hence, the uvrABC endonuclease network would not be able to identify and excise the 12 nucleotides of all genes consisting of the pyrimidine dimers. This leads to reduced colony growth as prokaryotic (K12 E. coli) cells cannot reproduce due to the insufficient production of proteins and enzymes such as DNA polymerase, DNA helicase, uvrABC, ligase, etc. Without the synthesis of these proteins and enzymes, DNA is unable to be transcribed, translated, replicated, or repaired (NER). When comparing “15 seconds” with the control samples, it is clear that the nucleotide excision repair mechanism was actively able to keep up with the minimal UV-C-induced damage within the DNA double helix of K12 E. coli. This is evident since the colonies in the “15 seconds” plates covered an average area of 47.83 cm² and the growth in the control plates covered an average area of 50.33 cm². Hence, when E. coli was exposed to UV-C for 15 seconds, it was still able to undergo binary fission allowing it to experience a growth similar to the control samples. However, when comparing “30 and 60 seconds'' with the control samples, the bacterial growth is significantly reduced. This is demonstrated in the results, as K12 E. coli was exposed to 30 seconds of UV-C when colonies over the 3 agar plates covered an average area of 29.67 cm², and the growth over “60” agar plates covered an average area of 8.5 cm² when exposed to UV-C for 60 seconds. This is because there would have been excessive distortions in the DNA double helix due to recurring cyclobutane pyrimidine dimers and 6-4 photoproducts. Hence, the nucleotide excision repair mechanism would experience a decreased capacity to keep up with these damages. This leads to several codons in genes potentially being misread by ribosomes or RNA polymerase, resulting in inaccurate protein production due to faulty mRNA coding for incorrect amino acids forming invalid polypeptide chains. Without proteins, prokaryotes may be unable to perform functions such as semi-conservative DNA replication or nucleotide excision repair (NER), potentially leading to cell death due to the absence of binary fission. Binary fission cannot occur in this scenario as there is no septum (cell wall) forming since the parent cell cannot partition a DNA molecule in both daughter cells. Finally, when comparing “120 and 300 seconds” with the control samples, the K12 E. coli growth is completely inactivated at a UV-C exposure of 300 seconds as no colonies or streaks were observed in any agar plates. However, minimal colony growth occurred over 2 agar plates, covering an average area of 0.33 cm² at a UV-C exposure of 120 seconds. However, at 300 seconds of exposure, the NER mechanism was completely unable to keep up with the pyrimidine dimers present in the DNA of K12 E. coli. Hence, processes of DNA replication and gene expression were completely terminated resulting in the absolute absence of binary fission.
A single outlier was observed within the recorded data. On the 3rd “60” nutrient agar plate, a significant amount of K12 E. coli colony growth was recorded with a total covered area of 14.5 cm². However, this concentrated growth only occurred in approximately one-half of the nutrient agar plate. This is abnormal because all the other “60” plates experienced very limited and scattered colony growth, expected at this duration of exposure. The most probable cause of this result was that one half of the nutrient agar plate with more dense colony growth was unintentionally positioned so that it was directed further from the UV-C source. Therefore, the UV-C intensity in this one-half of the plate was significantly less when compared to the other “60” plates. This allowed for greater colony growth due to the reduced presence of UV-C induced cyclobutane pyrimidine dimers and 6-4 photoproducts in that half, leading to the minimal presence of binary fission because the nucleotide excision repair mechanism could still counteract this damage to an extent. Another potential cause of this outlier could have been due to cross-contamination of foreign bacteria on the nutrient agar plate during the inoculation process. Although we wiped all the surfaces to keep the experiment environment as sterile as possible, bacteria other than K12 E. coli could have survived and eventually been transferred onto the plate.
Conclusion
- According to the data collected from this experiment, our hypothesis stating that 60 seconds would be the minimum duration to fully inactivate K12 Escherichia coli growth was proven incorrect. This is evident in the data, as the K12 Escherichia coli growth covered an average area of 8.5 cm² when exposed to UV-C for 60 seconds. Even after considering the outlier that recorded a total K12 Escherichia coli growth of 14.5 cm², agar plates 1 and 2 still collected a total growth covering 1 cm² and 1.5 cm².
- The reason behind the above occurrence is that when K12 Escherichia coli was exposed to UV-C radiation for 60 seconds, it was still able to experience limited binary fission due to minimal semi-conservative DNA replication. This is because the nucleotide excision repair mechanism was still able to counteract small amounts of cyclobutane pyrimidine dimers and 6-4 photoproducts.
- The minimum duration of UV-C exposure required to completely inactivate the cultivation of K12 Escherichia coli is 300 seconds.
Application
Application:
- Food and Meat Processing Companies: Numerous bacteria including E. coli are commonly found in meat. Therefore, stakeholders in this industry can use this data to determine the minimum duration of UV-C radiation required to completely terminate bacterial growth, leading to the enhanced safety of meat production.
- Households: Many households use UV-C to disinfect small areas in their house like bedrooms and closets. Homeowners can use the data from this experiment to determine how long they should apply UV-C to these areas in order to effectively remove all bacteria, ensuring a safer environment, especially during the COVID-19 pandemic, where risks of bacterial contamination are elevated.
- Water Treatment Facilities: Water treatment processes to remove bacteria often utilize UV-C radiation. Based on this experiment, water treatment system manufacturers, and companies can decide how long water needs to be exposed to UV-C radiation in order to fully inactivate all bacterial growth.
- Medical Environments: Bacteria are often found in medical clinics due to patients with infections. Hence, several types of bacteria are found on medical equipment and tools. We can place these tools in a UV-C sterilizer, and based on the recorded data we can identify that it would take 300 seconds to entirely kill bacteria on these equipment.
- Applying UV-C for them minimum duration is required in order to save on electricity bills and prevent alterations to meat taste, smell, and colour
Future Extensions:
In a scenario within which there were no constraints on our access to resources, we would have tested UV-C radiation on the complete inactivation of different types of bacteria such as salmonella or staphylococcus aureus. We hypothesize that if we expose different types of bacteria to UV-C then they would share a similar inactivation rate to K12 Escherichia coli as they are all examples of prokaryotes.
To extend our project we could investigate the effectiveness of hydrogen peroxide gas plasma sterilization techniques in the disinfection of contaminated surfaces. Therefore, we can explore the possibility of different bacterial inactivation techniques that could be more efficient compared to traditional methods. Other applications from this extension could also be incorporated in food packaging as hydrogen peroxide gas plasma sterilization techniques could possibly function as substitutes for preservatives.
Sources Of Error
- During the inoculation of the agar plates, despite our best efforts to keep the swabbing method consistent, some variation in patterns was inevitable, as we could not keep it the exact same every time.
- During the inoculation and radiation exposure processes, nitrile gloves were worn to ensure no contamination of the nutrient agar would occur. However, it is still possible that during these processes, some nutrient agar plates experienced cross-contamination with foreign bacteria.
- As stated above, the third “60” nutrient agar plate was most likely placed further from the UV-C source, thus allowing greater colony growth in comparison to the other nutrient agar plates.
- For this project, we did not include more durations (time intervals). Thus, it is possible that our results may not be as accurate as they could be.
Citations
Bandoim, L. (2019, May 21). Gene Expression in Prokaryotes. Sciencing. Retrieved November 15, 2023, from https://sciencing.com/gene-expression-in-prokaryotes-13717692.html
Bandoim, L. (2019, May 21). Prokaryotic Cells: Definition, Structure, Function (with Examples). Sciencing. Retrieved November 15, 2023, from https://sciencing.com/prokaryotic-cells-definition-structure-function-with-examples-13717657.html
Buckley, G. (2017, September 7). Nucleic Acid - Definition, Function and Examples. Biology Dictionary. Retrieved November 15, 2023, from https://biologydictionary.net/nucleic-acid/
Carter, S. (n.d.). Prokaryotic Transcription and Translation | Biology for Majors I. Lumen Learning. Retrieved November 15, 2023, from https://courses.lumenlearning.com/suny-wmopen-biology1/chapter/prokaryotic-transcription-and-translation/
Christensen, S. (2011, July 8). Amino Acids to Produce Collagen. Healthfully. Retrieved November 15, 2023, from https://healthfully.com/amino-acids-to-produce-collagen-7537871.html
Cotoia, A. (2020, June 1). DNA Replication - The Definitive Guide. Biology Dictionary. Retrieved January 12, 2024, from https://biologydictionary.net/dna-replication/
Cyclobutane. (n.d.). Wikipedia. Retrieved November 15, 2023, from https://en.wikipedia.org/wiki/Cyclobutane
Derworiz, C. (2023, December 31). Government panel probes failures in food safety leading to Calgary's E. Coli outbreak. CBC. Retrieved January 13, 2024, from https://www.cbc.ca/news/canada/calgary/e-coli-outbreak-calgary-looking-back-1.7071907
DNA. (n.d.). Wikipedia. Retrieved November 15, 2023, from https://en.wikipedia.org/wiki/DNA
DNA damage (naturally occurring) (n.d.) Retrieved November 15, 2023 from https://en.wikipedia.org/wiki/DNA_damage_%28naturally_occurring%29
DNA Definition & Usage Examples. (n.d.). Dictionary.com. Retrieved November 15, 2023, from https://www.dictionary.com/browse/DNA
DNA proofreading and repair (article). (n.d.). Khan Academy. Retrieved January 8, 2024, from https://www.khanacademy.org/science/biology/dna-as-the-genetic-material/dna-replication/a/dna-proofreading-and-repair
DNA replication - Replication of DNA - Higher Biology Revision. (n.d.). BBC. Retrieved November 15, 2023, from https://www.bbc.co.uk/bitesize/guides/zrwhrj6/revision/2
Effect of UV light on food quality and safety. (n.d.). ResearchGate. Retrieved December 18, 2023, from https://www.researchgate.net/publication/338474433_Effect_of_UV_light_on_food_quality_and_safety
Genetics - Medical Microbiology. (n.d.). NCBI. Retrieved November 15, 2023, from https://www.ncbi.nlm.nih.gov/books/NBK7908/
Gibson, K. (2018, September 26). Cargill ground beef recall after E. coli outbreak kills 1, sickens 17. CBS News. Retrieved January 13, 2024, from https://www.cbsnews.com/news/cargill-ground-beef-recall-after-e-coli-outbreak-that-has-killed-1-person-sickened-17/
Gillespie, C. (2018, April 13). Colony Characteristics of E.Coli. Sciencing. Retrieved November 15, 2023, from https://sciencing.com/colony-characteristics-ecoli-8507841.html
Helicase. (n.d.). Wikipedia. Retrieved November 15, 2023, from https://en.wikipedia.org/wiki/Helicase
Latham, K. (2021, January 9). DNA vs. RNA. Biology Dictionary. Retrieved November 15, 2023, from https://biologydictionary.net/dna-vs-rna/
List of strains of Escherichia coli. (n.d.). Wikipedia. Retrieved November 15, 2023, from https://en.wikipedia.org/wiki/List_of_strains_of_Escherichia_coli
Meštrović, T., & Surat, P. (2021, January 27). A Guide to Understanding Gene Expression. AZoLifeSciences.com. Retrieved January 6, 2024, from https://www.azolifesciences.com/article/A-Guide-to-Understanding-Gene-Expression.aspx
The need for DNA replication - Replication of DNA - Higher Biology Revision. (n.d.). BBC. Retrieved November 15, 2023, from https://www.bbc.co.uk/bitesize/guides/zrwhrj6/revision/1
*30 points* The diagram shows one step in the process of protein synthesis. Which step is shown? - brainly.com. (2017, June 1). Brainly. Retrieved January 11, 2024, from https://brainly.com/question/3955033
Nucleotide excision repair. (n.d.). Wikipedia. Retrieved November 15, 2023, from https://en.wikipedia.org/wiki/Nucleotide_excision_repair
Nutrition of Bacteria - Structure of Bacteria. (2021, September 22). DesktopClass.com. Retrieved January 6, 2024, from https://www.desktopclass.com/biology/structure-of-bacteria-part-4-f-sc-biology-chapter-6/
Protecting your eyes from the sun’s UV light. (2022, July 5). National Eye Institute. Retrieved January 6, 2024, from https://www.nei.nih.gov/about/news-and-events/news/protecting-your-eyes-suns-uv-light
Pyrimidine dimer. (n.d.). Wikipedia. Retrieved November 15, 2023, from https://en.wikipedia.org/wiki/Pyrimidine_dimer
Pyrimidine Dimers: An Overview. (2021). Science Direct. Retrieved November 13, 2023, from https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/pyrimidine-dimer
Pyrimidine - The Definitive Guide. (2017, August 6). Biology Dictionary. Retrieved January 11, 2024, from https://biologydictionary.net/pyrimidine/
RNA Definition & Usage Examples. (n.d.). Dictionary.com. Retrieved November 15, 2023, from https://www.dictionary.com/browse/RNA
Structural Biochemistry/Organic Chemistry/Nucleic Acids (DNA). (n.d.). Wikibooks. Retrieved November 27, 2023, from https://en.wikibooks.org/wiki/Structural_Biochemistry/Organic_Chemistry/Nucleic_Acids_(DNA)
Ultraviolet - Wikipedia. (2021, September 22). Retrieved November 15, 2023, from https://en.wikipedia.org/w/index.php?title=Ultraviolet&oldid=873163862
What are the monomers of DNA and RNA? (n.d.). Homework.Study.com. Retrieved January 11, 2024, from https://homework.study.com/explanation/what-are-the-monomers-of-dna-and-rna.html
What are the structure of RNA and DNA and how one might be used to create the other? (2018, June 17). Socratic. Retrieved January 11, 2024, from https://socratic.org/questions/what-are-the-structure-of-rna-and-dna-and-how-one-might-be-used-to-create-the-ot
What is E coli and what are the symptoms of infection? (2018, September 12). The Independent. Retrieved February 29, 2024, from https://www.independent.co.uk/life-style/health-and-families/e-coli-bacteria-symptoms-infection-egypt-outbreak-treatment-explained-a8525486.html
Acknowledgement
We would like to express our deepest gratitude to our parents and teachers for their unwavering support throughout the course of this project. We would also like to give special acknowledgement to Ms. Kerry Ball, a junior high school teacher with a background in microbiology, for reviewing the related documents and providing feedback. Lastly, a great thanks goes to Tejas Oza with a Ph.D. in microbiology, for reviewing our lab report and providing helpful suggestions. The support of these people was invaluable to the success and progression of this project.