How can specific DNA methylation patterns enhance the early detection of leukemia?
Grade 10
Presentation
No video provided
Problem
Leukemia is a devastating disease that affects millions of people worldwide, disrupting lives and placing an immense burden on patients and their families. The inspiration behind my project stems from the urgent need for more precise, early detection methods that can improve outcomes and develop a more efficient healthcare system. DNA methylation patterns play a crucial role in leukemia development, yet its potential as a diagnostic tool remains underutilized. By studying methylation patterns in key leukemia-associated genes, I aim to contribute to the growing body of research focused on detecting the disease at its earliest stages. With leukemia impacting children (accounting for 25% of all childhood cancers) and adults globally, advancements in early detection and targeted treatments could save countless lives and provide hope for those battling this condition.
Method
Methodology
Step 1: Research Leukemia Types and Severity
-
Conducted literature reviews using PubMed, Google Scholar, NCBI, and TCGA to analyze the main leukemia types (AML, ALL).
-
Studied their genetic, epigenetic, and clinical characteristics from multiple peer-reviewed sources to ensure a well-rounded understanding of disease progression and severity.
Step 2: Analyze Current Leukemia Diagnosis Methods and Limitations
-
Reviewed existing leukemia diagnostic methods (CBC, bone marrow biopsy, flow cytometry) through research papers and clinical studies from PubMed, NCBI, and GEO.
-
Identified commonly reported inefficiencies, such as:
-
CBC lacks specificity in distinguishing leukemia from other blood disorders.
-
Bone marrow biopsies are invasive and costly.
-
Flow cytometry requires specialized expertise and equipment.
-
-
Cross-verified findings across multiple sources to ensure accuracy and completeness of the analysis.
Step 3: Study Epigenetic Biomarkers and DNA Methylation Patterns
-
Researched epigenetic biomarkers, particularly DNA methylation patterns, and its role in gene regulation.
-
Studied how DNA methylation pattern changes can act as potential indicators for leukemia.
-
Focused on key leukemia-associated methylation markers and their success in identifying the cancer cells.
Step 4: Detect Leukemia-Specific DNA Methylation Patterns
-
Gathered leukemia-related DNA methylation data from PubMed, Google Scholar, NCBI, TCGA, and GEO.
-
Cross-verified findings across multiple sources to confirm reliability and reduce inconsistencies.
-
Conducted additional research on methods used to extract and analyze methylation patterns, including:
-
Bisulfite sequencing
-
Methylation-specific PCR (MSP)
-
Computational tools like DMRcaller and MethyKit
-
Step 5: Assess the Efficiency of DNA Methylation Patterns for Leukemia Diagnosis
-
Evaluated the sensitivity and specificity of DNA methylation-based diagnosis using published studies.
-
Explored computational models to classify leukemia subtypes based on methylation signatures.
-
Justified whether the current diagnosis process or the DNA Methylation Pattern identification is more precise, non-invasive and effective.
Research
Leukemia Types :
| Causes | Symptoms | Diagnosis | Treatment | |
|
Acute Lymphoblastic Leukemia (ALL) The most common type in children and adolescents (children under the age of 4, and males). Originates in the lymphoid stem cells of the immune system and affects the cells that produce lymphocytes, which fight infection. ALL can develop anywhere in the body where blood travels. |
|
*Can be vague and resemble symptoms of the flu and other common illnesses*
|
|
*Depends on the patient's age, general health, and sub-type of ALL*
|
|
Acute Myeloid Leukemia (AML) More common and aggressive in adults. Develops from the myeloid cells that produce red blood cells, white blood cells, and platelets.
|
Occurs when there's a DNA mutation in the stem cells of the bone marrow. The mutation causes the stem cells to produce too many immature white blood cells, called blast cells, which crowd out healthy blood cells. *unknown exact causes of AML
|
|
|
*Depends on the patient's age, general health, and sub-type of AML
|
*Prevention : (No known way to prevent most cases of leukemia)
- Avoiding tobacco
- Healthy diet - rich in fruits and vegetables and low in processed meats.
- Maintaining healthy weight
- Limiting chemical exposure (benzene and formaldehyde)
- Staying physically active - proven to reduce risk of cancer in general
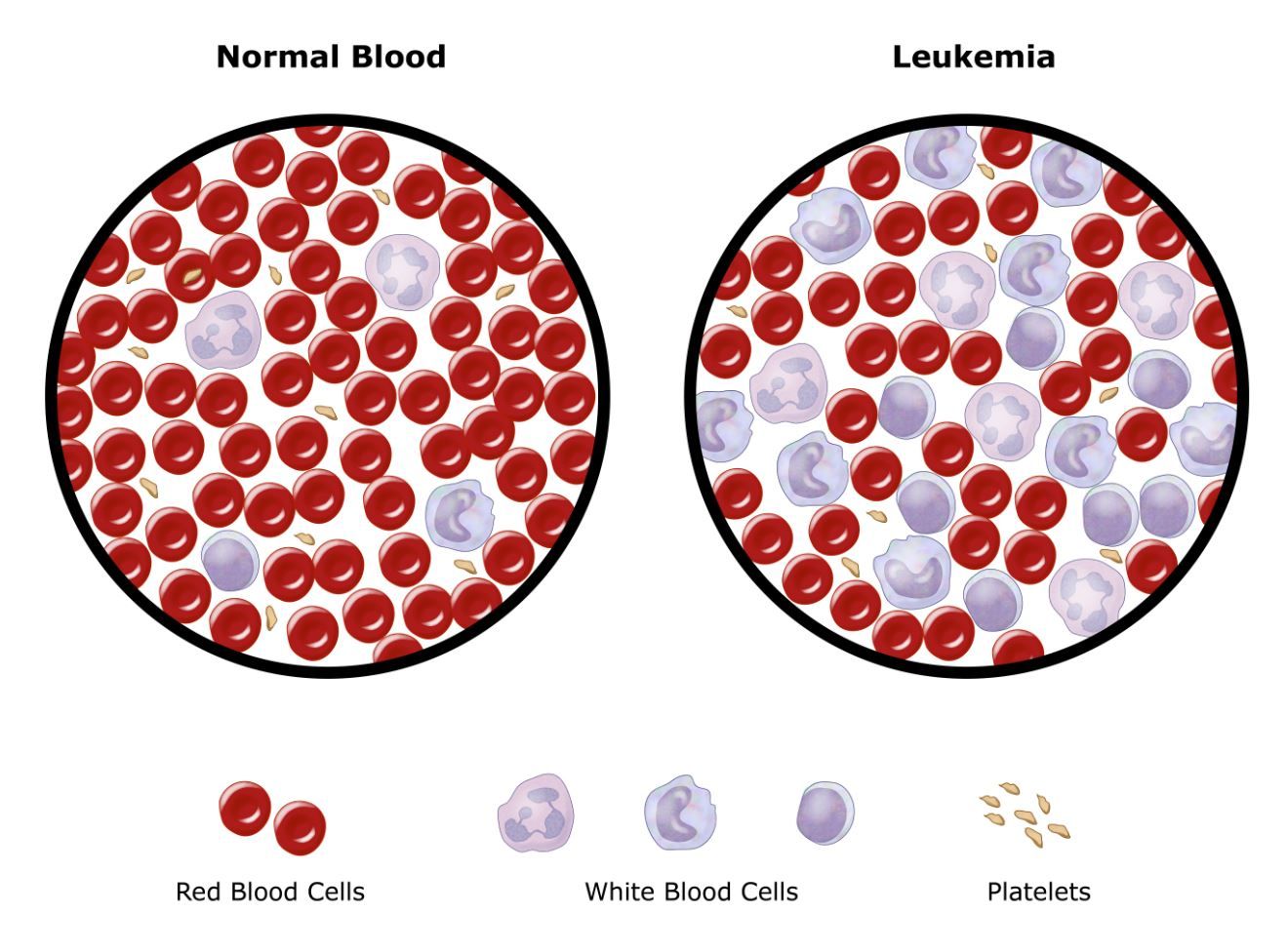
normal cells, overcrowding the bone marrowand preventing the
normal cells from functioning properly.
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Current Healthcare System and Failures (Canada Specific) :
In the Canadian healthcare system, the Complete Blood Count (CBC) is the most common initial test used to detect leukemia. It measures different blood cell levels and can indicate abnormalities, such as elevated white blood cell counts or anemia, which may suggest leukemia. However, CBC lacks specificity and often fails to detect leukemia in its early stages. For a definitive diagnosis, a bone marrow biopsy is required, where a sample is examined for leukemia cells. While effective, this procedure is invasive and unsuitable for routine screening.
Additional tests, such as immunophenotyping and cytogenetic analyses, help identify leukemia subtypes and guide treatment. However, these methods are costly, time-consuming, and only performed once leukemia is already suspected. As a result, diagnoses are often delayed, particularly in individuals without severe symptoms. By the time leukemia is detected, it may already be in an advanced stage, limiting treatment options and reducing survival rates.
Limitations of Current Leukemia Detection Methods in the Canadian Healthcare System :
| Method | Why It’s Used | Why It’s Inefficient |
| Complete Blood Count (CBC) | First-line test to detect abnormal blood cell levels that may indicate leukemia. | Cannot detect early-stage leukemia before symptoms appear and doesn’t specify leukemia subtype. Further testing is always required. |
| Bone Marrow Biopsy | Confirms leukemia diagnosis by analyzing bone marrow cells for cancer presence. | Highly invasive and painful, requiring anesthesia and recovery time. It is not ideal for routine screening or early detection. |
| Immunophenotyping (Flow Cytometry) | Identifies leukemia subtype by analyzing proteins on blood or bone marrow cells. | Expensive and time-consuming, often requiring specialized lab equipment and expertise, making it impractical for widespread early detection. |
| Cytogenetic Testing (Karyotyping/FISH/PCR) | Detects genetic mutations linked to specific leukemia types for targeted treatment. | Requires advanced lab techniques, making it unavailable in some smaller medical centers. Also, it does not help in early detection—only in classifying leukemia after it’s diagnosed. |
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Epigenetic Biomarkers : Epigenetic biomarkers are chemical modifications to DNA, histones, or RNA that regulate gene expression without altering the underlying genetic sequence. These biomarkers serve as indicators of disease states, environmental exposures, aging, and cancer progression and can be used for early detection, prognosis, and treatment monitoring.
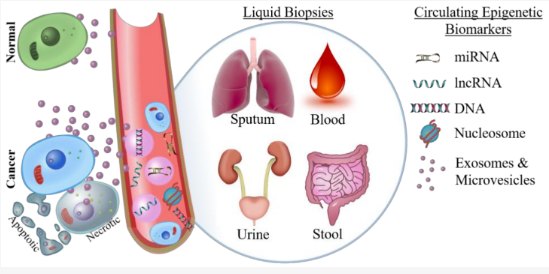
Location :
- DNA Methylation in Genes (Inside the Nucleus) - CpG islands are regions in DNA that are rich in cytosine (C) and guanine (G) pairs, and they play a crucial role in gene regulation. When these regions become methylated (chemically modified with a methyl group -CH3), they can either activate or deactivate genes. The methylation of CpG islands is a regulatory mechanism for gene expression, and when it becomes disrupted, it can contribute to diseases like cancer by silencing important protective genes.
- Histone Modifications (Inside the Nucleus, in Chromatin) - Histones are proteins that help package DNA into a compact structure called chromatin. They can be chemically modified by adding acetyl, methyl, or phosphate groups, which either loosen or tighten DNA packaging. This controls how easily genes can be turned on or off.
- Non-Coding RNAs (Floating in the Cell and Bloodstream) - Some RNA molecules, like microRNAs (miRNAs), regulate gene expression without the need to produce proteins. Certain miRNAs are considered cancer markers because their levels fluctuate in cancer patients.
- Circulating Cell-Free DNA (cfDNA) and Circulating Tumor DNA (ctDNA) (Found in Blood & Other Bodily Fluids) - Cells shed small fragments of DNA into the bloodstream when they die. In cancer patients, tumor cells release circulating tumor DNA (ctDNA), which carries epigenetic changes. Detecting methylation changes in ctDNA can help with early cancer detection using liquid biopsies.
DNA Methylation Patterns : DNA methylation is an epigenetic mechanism that modifies DNA without altering its underlying sequence. Essentially, it is the addition of a methyl group (CH₃) to the DNA molecule at the 5 position of the cytosine ring, resulting in the formation of 5-methylcytosine (5mC). These methylation patterns determine gene expression by influencing how the genes are read and transcribed. They can recruit proteins important for gene repression or inhibit the binding of transcription factors to DNA, leading to the gene's activity regulation. These marks are good indicators because they are stable and can be preserved in biological samples like blood, saliva, and tissue. Cancer is detected by specific, abnormal methylation patterns found in certain genes.
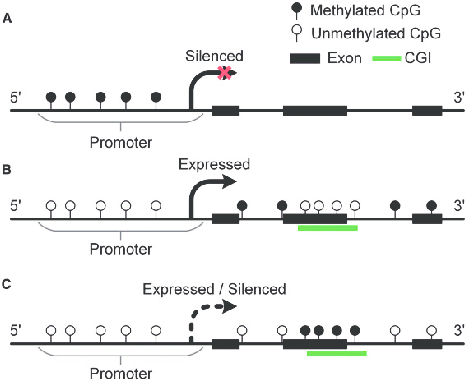
Extraction Methods : *Easiest and most common methods*
{DNA extraction kit: DMRcaller, MethyKit, or ChAMP (R packages)}
- Blood Sample: Isolate DNA from blood cells
- Tissue Sample: Homogenize the tissue
- Saliva Sample
- Buccal Swab: Collect cells from the cheek
- Plasma/Serum: Extract circulating DNA using specialized kits for cfDNA (circulating free DNA)
DNA Methylation Analysis :
- Bisulfite Conversion: The first step is to modify the DNA so that unmethylated cytosines are converted to uracils, while methylated cytosines stay the same. This allows for the distinction between methylated and unmethylated DNA.
- Methylation-Specific PCR (MSP): After bisulfite conversion, MSP is used to specifically amplify and analyze either methylated or unmethylated DNA regions. It helps detect methylation in specific cancer-related genes.
- Next-Generation Sequencing (NGS): For a more comprehensive view, NGS can be used after bisulfite treatment to sequence the entire genome or targeted regions, providing a detailed map of DNA methylation changes across many cancer-related genes.
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Comparison of DNA Methylation Pattern Analysis vs. CBC in Leukemia Detection :
| Feature | DNA Methylation Analysis | Complete Blood Count (CBC) |
| What It Measures | Epigenetic modifications (methylation changes) in leukemia-associated genes. | Number and proportion of different blood cells (white blood cells, red blood cells, platelets). |
| Detection Mechanism | Identifies leukemia-specific epigenetic changes (abnormal gene silencing or activation due to methylation). | Detects abnormal cell counts (e.g., high white blood cell count, low platelets, anemia) but does not explain why they are abnormal. |
| Sensitivity & Specificity | Can detect molecular changes before leukemia symptoms appear, offering potential for very early detection. | Often identifies leukemia after symptoms develop, when abnormal blood counts are already present. |
| Type of Data Analyzed | DNA from blood or bone marrow samples, analyzed for methylation patterns in genes linked to leukemia. | Quantitative count of blood cells under a microscope or via automated machines. |
| Role in Diagnosis | Can help classify different leukemia subtypes (e.g., AML, ALL) based on epigenetic profiles, improving targeted treatments. | Suggests leukemia only if blood cell counts are significantly abnormal, requiring further tests like bone marrow biopsy. |
| Long-Term Potential | May lead to non-invasive, blood-based early screening tools for high-risk individuals (e.g., those with family history or prior cancers). | Used mainly for routine monitoring rather than early-stage detection. |
Data
Leukemia Specific Methylation Patterns :
|
Gene Type |
Gene |
Location |
Methylation Pattern |
|
Tumor Suppressors |
CDKN2A (p16INK4a) |
Chromosome 9p21 |
silencing in AML, ALL |
|
PTEN |
Chromosome 10q23.31 |
silencing in AML |
|
|
DAPK1 |
Chromosome 9q34.1 |
silencing in AML, ALL |
|
|
Oncogenes |
HOXA Cluster |
Chromosome 7p15-p14 |
overexpression in leukemia |
|
WT1 |
Chromosome 11p13 |
silencing in AML, ALL |
|
|
Epigenetic Regulators |
TET2 |
Chromosome 4q24 |
silencing in AML, MDS |
|
DNMT3A |
Chromosome 2p23.3 |
silencing in AML and other cancers |
|
|
IDH1/IDH2 |
Chromosome 2q34 (IDH1), Chromosome 15q26.1 (IDH2) |
expression in AML and other hematological cancers |
|
|
Immune Pathway Genes |
SOCS1 |
Chromosome 16p13.3 |
silencing in AML, CLL |
|
FAS |
Chromosome 10q24.1 |
silencing in AML, ALL |
Conclusion
Leukemia often develops gradually, and traditional methods like the Complete Blood Count (CBC) can only detect it once significant changes in blood cell numbers occur. This means leukemia is often diagnosed after symptoms appear, significantly delaying early intervention.
In contrast, DNA methylation analysis offers a much more advanced approach. Instead of waiting for abnormal cell counts, this method looks at epigenetic changes—modifications in DNA that regulate gene expression. These changes often occur before leukemia fully develops, making this method an excellent tool for early detection. Additionally, different subtypes of leukemia (e.g., AML, ALL, CLL) have unique methylation signatures, allowing for more precise classification and personalized treatment.
Another advantage is its potential use as a non-invasive blood test. High-risk individuals, such as those with a family history of leukemia or previous cancer treatments, could undergo routine methylation screening to catch leukemia before traditional methods detect it. This could revolutionize early diagnosis and patient outcomes.
Citations
Sources : PubMed, Google Scholar, NCBI, The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO)
| Leukemia |
- Snodgrass, Rayven, et al. “Incidence of Acute Lymphocytic Leukemia in Calgary, Alberta, Canada: A Retrospective Cohort Study.” BMC Research Notes, vol. 11, no. 1, Feb. 2018, https://doi.org/10.1186/s13104-018-3225-9. - Public Health Agency of Canada. “Cancer in Adolescents in Canada (15-19 Years).” Canada.ca, 9 July 2012, www.canada.ca/en/public-health/services/chronic-diseases/cancer/cancer-adolescents-canada-15-19-years.html. - Acute Lymphoblastic Leukemia (ALL): MedlinePlus Medical Encyclopedia. medlineplus.gov/ency/article/000541.htm#:~:text=Causes&text=Most%20of%20the%20time%2C%20no,Toxins%2C%20such%20as%20benzene. - “Signs and Symptoms of Acute Lymphocytic Leukemia (ALL).” American Cancer Society, www.cancer.org/cancer/types/acute-lymphocytic-leukemia/detection-diagnosis-staging/signs-symptoms.html. - “Typical Treatment of Acute Lymphocytic Leukemia (ALL).” American Cancer Society, www.cancer.org/cancer/types/acute-lymphocytic-leukemia/treating/typical-treatment.html#:~:text=As%20the%20leukemia%20grows%20in,may%20be%20treated%20with%20antibiotics. - Du Cancer, Canadian Cancer Society /. Société Canadienne. “Classification of Acute Lymphoblastic Leukemia.” Canadian Cancer Society, cancer.ca/en/cancer-information/cancer-types/acute-lymphoblastic-leukemia-all/what-is-acute-lymphoblastic-leukemia/classification-of-acute-lymphocytic-leukemia#:~:text=About%201%20in%204%20adults,more%20common%20in%20older%20people. - “Typical Treatment of Acute Myeloid Leukemia (Except APL).” American Cancer Society, www.cancer.org/cancer/types/acute-myeloid-leukemia/treating/typical-treatment-of-aml.html#:~:text=People%20typically%20need%20to%20stay,low%20for%20a%20few%20weeks. - “Acute Myeloid Leukemia Treatment.” Cancer.gov, 28 Feb. 2025, www.cancer.gov/types/leukemia/patient/adult-aml-treatment-pdq. |
| Current System |
- Adamska, Monika, et al. “Clinical Outcomes of Therapy-related Acute Myeloid Leukemia: Over 20 Years Long Single Center Retrospective Analysis.” Polskie Archiwum Medycyny Wewnętrznej, Sept. 2022, https://doi.org/10.20452/pamw.16344. - Mph, Carrie Madormo Rn. “How Is Leukemia Diagnosed?” Health, 10 Jan. 2025, www.health.com/testing-for-leukemia-diagnosis-8740204?utm. - https://www.cancercenter.com/cancer-types/leukemia/diagnosis-and-detection - Canada, Leukemia &. Lymphoma Society Of. “Understanding Lab Results.” The Leukemia & Lymphoma Society of Canada, www.bloodcancers.ca/understanding-lab-results. - Guideline Resource Unit. “Acute Myeloid Leukemia.” Guideline Resource Unit, Apr. 2024, p. 2. www.albertahealthservices.ca/assets/info/hp/cancer/if-hp-cancer-guide-lyhe006-aml.pdf. - Wikipedia contributors. “Complete Blood Count.” Wikipedia, 27 Feb. 2025, en.wikipedia.org/wiki/Complete_blood_count?utm. |
| Epigenetic Biomarkers |
- Chen, Xi, et al. “Pan-Cancer Epigenetic Biomarker Selection From Blood Samples Using SAS.” arXiv.org, 21 Dec. 2018, arxiv.org/abs/1812.09203. - Esteller, Manel. “Epigenetics in Cancer.” New England Journal of Medicine, vol. 358, no. 11, Mar. 2008, pp. 1148–59. https://doi.org/10.1056/nejmra072067. - Laird, Peter W. “Principles and Challenges of Genome-wide DNA Methylation Analysis.” Nature Reviews Genetics, vol. 11, no. 3, Feb. 2010, pp. 191–203. https://doi.org/10.1038/nrg2732. - Figueroa, M. E., et al. (2010). "DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia." Cancer Cell, 17(1), 13-27. www.cancer.gov/ccg/research/genome-sequencing/tcga. - Bannister, Andrew J., and Tony Kouzarides. “Regulation of Chromatin by Histone Modifications.” Cell Research, vol. 21, no. 3, Feb. 2011, pp. 381–95. https://doi.org/10.1038/cr.2011.22. - Calin, George A., and Carlo M. Croce. “MicroRNA Signatures in Human Cancers.” Nature Reviews. Cancer, vol. 6, no. 11, Oct. 2006, pp. 857–66. https://doi.org/10.1038/nrc1997. - Siravegna, Giulia, et al. “Integrating Liquid Biopsies Into the Management of Cancer.” Nature Reviews Clinical Oncology, vol. 14, no. 9, Mar. 2017, pp. 531–48. https://doi.org/10.1038/nrclinonc.2017.14. |
| DNA Methylation Patterns |
- Moore, Lisa D., et al. “DNA Methylation and Its Basic Function.” Neuropsychopharmacology, vol. 38, no. 1, July 2012, pp. 23–38. https://doi.org/10.1038/npp.2012.112. - “DNA Methylation.” Wikipedia, 17 Feb. 2025, en.wikipedia.org/wiki/DNA_methylation. - Nordlund, Jessica, et al. “DNA Methylation-based Subtype Prediction for Pediatric Acute Lymphoblastic Leukemia.” Clinical Epigenetics, vol. 7, no. 1, Feb. 2015, https://doi.org/10.1186/s13148-014-0039-z. - “DNA Methylation-based Subtype Prediction for Pediatric Acute Lymphoblastic Leukemia.” Clinical Epigenetics, vol. 7, no. 1, Feb. 2015, https://doi.org/10.1186/s13148-014-0039-z. |
Photos
- https://orthoinfo.aaos.org/en/diseases--conditions/leukemia/
- https://www.cancer.gov/publications/dictionaries/cancer-terms/def/blood-stem-cell
- https://www.researchgate.net/figure/Cartoon-graph-of-the-correlations-between-DNA-methylation-and-gene-expression-A_fig1_301610625https://www.mdpi.com/2072-6694/10/4/101
- https://www.researchgate.net/figure/Cartoon-graph-of-the-correlations-between-DNA-methylation-and-gene-expression-A_fig1_301610625
Videoes
- https://www.youtube.com/watch?v=Z3B-AaqjyjE
- https://www.youtube.com/watch?v=j0ruBn1XM24&t=405s
- https://www.youtube.com/watch?v=6HwptTI0Pxw
- https://www.youtube.com/watch?v=UUM7HiFkDd4
- https://www.youtube.com/watch?v=_NBo-GZDKOM
- https://www.youtube.com/watch?v=dhR8bjD8D2Q
- https://www.youtube.com/watch?v=QBpgU34jxYM
Acknowledgement
I am grateful to many people who have joined me in this endeavor of another great year spent fascinating over science and endless discoveries! First and foremost, I am profoundly grateful to my parents - Hira and Uma Rawat, for their unwavering assistance, continuous encouragement, and boundless sacrifices throughout this project. Their support, both emotionally and materially, has been the cornerstone of my project's success. Their relentless devotion, whether it be rides to gather supplies, or a constant supply of snacks during work periods, has enriched the depth and clarity of my project, enabling me to find and understand data effectively. Their belief in my abilities has been a constant source of motivation, driving me to strive for excellence. I am truly blessed to have such dedicated and supportive parents by my side, and I thank them from the bottom of my heart for everything they have done. Next, I would like to express my biggest thanks to my science fair coordinators and amazing teachers - Ms. O'Keefe, and Ms.Jonhson for their constant presence and unwavering support throughout this process. Their persistent push of encouragement and participation in our progress is what has kept me on track, along with their valuable suggestions. I am truly grateful for all they have helped us with and for making science fair a memorable journey! Lastly, but not the least, I extend my thanks to all those who directly or indirectly contributed to the realization of this project, as their collective efforts have undoubtedly enhanced its quality.

